Crystal structure of complete rhinovirus RNA polymerase suggests front loading of protein primer
- PMID: 15596823
- PMCID: PMC538717
- DOI: 10.1128/JVI.79.1.277-288.2005
Crystal structure of complete rhinovirus RNA polymerase suggests front loading of protein primer
Abstract
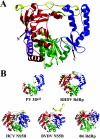
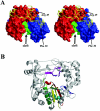
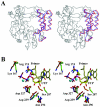
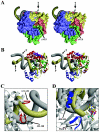
Picornaviruses utilize virally encoded RNA polymerase and a uridylylated protein primer to ensure replication of the entire viral genome. The molecular details of this mechanism are not well understood due to the lack of structural information. We report the crystal structure of human rhinovirus 16 3D RNA-dependent RNA polymerase (HRV16 3Dpol) at a 2.4-A resolution, representing the first complete polymerase structure from the Picornaviridae family. HRV16 3Dpol shares the canonical features of other known polymerase structures and contains an N-terminal region that tethers the fingers and thumb subdomains, forming a completely encircled active site cavity which is accessible through a small tunnel on the backside of the molecule. The small thumb subdomain contributes to the formation of a large cleft on the front face of the polymerase which also leads to the active site. The cleft appears large enough to accommodate a template:primer duplex during RNA elongation or a protein primer during the uridylylation stage of replication initiation. Based on the structural features of HRV16 3Dpo1 and the catalytic mechanism known for all polymerases, a front-loading model for uridylylation is proposed.
Figures


Similar articles
-
The crystal structure of the RNA-dependent RNA polymerase from human rhinovirus: a dual function target for common cold antiviral therapy.Structure. 2004 Aug;12(8):1533-44. doi: 10.1016/j.str.2004.05.024. Structure. 2004. PMID: 15296746
-
RNA-dependent RNA polymerases from Flaviviridae.Curr Opin Struct Biol. 2009 Dec;19(6):746-51. doi: 10.1016/j.sbi.2009.10.015. Epub 2009 Nov 14. Curr Opin Struct Biol. 2009. PMID: 19914821 Review.
-
Structural insights into replication initiation and elongation processes by the FMDV RNA-dependent RNA polymerase.Curr Opin Struct Biol. 2009 Dec;19(6):752-8. doi: 10.1016/j.sbi.2009.10.016. Epub 2009 Nov 13. Curr Opin Struct Biol. 2009. PMID: 19914060 Review.
-
Nucleotide channel of RNA-dependent RNA polymerase used for intermolecular uridylylation of protein primer.J Mol Biol. 2006 Mar 24;357(2):665-75. doi: 10.1016/j.jmb.2005.12.044. Epub 2006 Jan 5. J Mol Biol. 2006. PMID: 16427083
-
The structure of a protein primer-polymerase complex in the initiation of genome replication.EMBO J. 2006 Feb 22;25(4):880-8. doi: 10.1038/sj.emboj.7600971. Epub 2006 Feb 2. EMBO J. 2006. PMID: 16456546 Free PMC article.
Cited by
-
The RNA template channel of the RNA-dependent RNA polymerase as a target for development of antiviral therapy of multiple genera within a virus family.PLoS Pathog. 2015 Mar 23;11(3):e1004733. doi: 10.1371/journal.ppat.1004733. eCollection 2015 Mar. PLoS Pathog. 2015. PMID: 25799064 Free PMC article.
-
Structural Analysis of Monomeric RNA-Dependent Polymerases: Evolutionary and Therapeutic Implications.PLoS One. 2015 Sep 23;10(9):e0139001. doi: 10.1371/journal.pone.0139001. eCollection 2015. PLoS One. 2015. PMID: 26397100 Free PMC article.
-
Fidelity Variants and RNA Quasispecies.Curr Top Microbiol Immunol. 2016;392:303-22. doi: 10.1007/82_2015_483. Curr Top Microbiol Immunol. 2016. PMID: 26499340 Free PMC article. Review.
-
Expanding knowledge of P3 proteins in the poliovirus lifecycle.Future Microbiol. 2010 Jun;5(6):867-81. doi: 10.2217/fmb.10.40. Future Microbiol. 2010. PMID: 20521933 Free PMC article. Review.
-
The crystal structure of a cardiovirus RNA-dependent RNA polymerase reveals an unusual conformation of the polymerase active site.J Virol. 2014 May;88(10):5595-607. doi: 10.1128/JVI.03502-13. Epub 2014 Mar 5. J Virol. 2014. PMID: 24600002 Free PMC article.
References
-
- Ago, H., T. Adachi, A. Yoshida, M. Yamamoto, N. Habuka, K. Yatsunami, and M. Miyano. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure Fold Des. 7:1417-1426. - PubMed
-
- Blessing, R. H., and G. D. Smith. 1999. Difference structure factor normalization for determining heavy-atom or anomalous scattering substructures. J. Appl. Cryst. 32:664-670.
-
- Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bamford, and D. I. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235-240. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

