Role for tumor necrosis factor alpha in murine cytomegalovirus transcriptional reactivation in latently infected lungs
- PMID: 15596827
- PMCID: PMC538715
- DOI: 10.1128/JVI.79.1.326-340.2005
Role for tumor necrosis factor alpha in murine cytomegalovirus transcriptional reactivation in latently infected lungs
Abstract
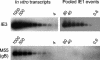
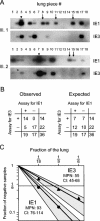
Interstitial pneumonia is a major clinical manifestation of primary or recurrent cytomegalovirus (CMV) infection in immunocompromised recipients of a bone marrow transplant. In a murine model, lungs were identified as a prominent site of CMV latency and recurrence. Pulmonary latency of murine CMV is characterized by high viral genome burden and a low incidence of variegated immediate-early (IE) gene expression, reflecting a sporadic activity of the major IE promoters (MIEPs) and enhancer. The enhancer-flanking promoters MIEP1/3 and MIEP2 are switched on and off during latency in a ratio of approximately 2:1. MIEP1/3 latency-associated activity generates the IE1 transcript of the ie1/3 transcription unit but not the alternative splicing product IE3 that encodes the essential transactivator of early gene expression. Splicing thus appeared to be an important checkpoint for maintenance of latency. In accordance with previous work of others, we show here that signaling by the proinflammatory cytokine tumor necrosis factor alpha (TNF-alpha) activates IE1/3 transcription in vivo. As an addition to current knowledge, Poisson distribution analysis revealed an increased incidence of IE1/3 transcriptional events as well as a higher amount of transcripts per event. Notably, TNF-alpha promoted the splicing to IE3 transcripts, but transcription did not proceed to the M55/gB early gene. Moreover, the activated transcriptional state induced by TNF-alpha did not predispose latently infected mice to a higher incidence of virus recurrence after hematoablative treatment. In conclusion, TNF-alpha is an important inductor of IE gene transcriptional reactivation, whereas early genes downstream in the viral replicative cycle appear to be the rate-limiting checkpoint(s) for virus recurrence.
Figures


Similar articles
-
Random, asynchronous, and asymmetric transcriptional activity of enhancer-flanking major immediate-early genes ie1/3 and ie2 during murine cytomegalovirus latency in the lungs.J Virol. 2001 Mar;75(6):2692-705. doi: 10.1128/JVI.75.6.2692-2705.2001. J Virol. 2001. PMID: 11222693 Free PMC article.
-
CD8 T cells control cytomegalovirus latency by epitope-specific sensing of transcriptional reactivation.J Virol. 2006 Nov;80(21):10436-56. doi: 10.1128/JVI.01248-06. Epub 2006 Aug 23. J Virol. 2006. PMID: 16928768 Free PMC article.
-
Focal transcriptional activity of murine cytomegalovirus during latency in the lungs.J Virol. 1999 Jan;73(1):482-94. doi: 10.1128/JVI.73.1.482-494.1999. J Virol. 1999. PMID: 9847354 Free PMC article.
-
Mouse models of cytomegalovirus latency: overview.J Clin Virol. 2002 Aug;25 Suppl 2:S23-36. doi: 10.1016/s1386-6532(02)00087-2. J Clin Virol. 2002. PMID: 12361754 Review.
-
A model for reactivation of CMV from latency.J Clin Virol. 2002 Aug;25 Suppl 2:S123-36. doi: 10.1016/s1386-6532(02)00088-4. J Clin Virol. 2002. PMID: 12361763 Review.
Cited by
-
Update on the management of ulcerative colitis.Curr Gastroenterol Rep. 2011 Oct;13(5):475-85. doi: 10.1007/s11894-011-0216-6. Curr Gastroenterol Rep. 2011. PMID: 21789495 Review.
-
Insufficient Antigen Presentation Due to Viral Immune Evasion Explains Lethal Cytomegalovirus Organ Disease After Allogeneic Hematopoietic Cell Transplantation.Front Cell Infect Microbiol. 2020 Apr 15;10:157. doi: 10.3389/fcimb.2020.00157. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32351904 Free PMC article.
-
The myeloid transcription factor GATA-2 regulates the viral UL144 gene during human cytomegalovirus latency in an isolate-specific manner.J Virol. 2013 Apr;87(8):4261-71. doi: 10.1128/JVI.03497-12. Epub 2013 Jan 30. J Virol. 2013. PMID: 23365437 Free PMC article.
-
Cytokine-Mediated Induction and Regulation of Tissue Damage During Cytomegalovirus Infection.Front Immunol. 2019 Jan 29;10:78. doi: 10.3389/fimmu.2019.00078. eCollection 2019. Front Immunol. 2019. PMID: 30761144 Free PMC article. Review.
-
Stochastic Episodes of Latent Cytomegalovirus Transcription Drive CD8 T-Cell "Memory Inflation" and Avoid Immune Evasion.Front Immunol. 2021 Apr 22;12:668885. doi: 10.3389/fimmu.2021.668885. eCollection 2021. Front Immunol. 2021. PMID: 33968074 Free PMC article.
References
-
- Adam, E., V. Quivy, F. Bex, A. Chariot, Y. Collette, C. Vanhulle, S. Schoonbroodt, V. Goffin, T. L. Nguyen, G. Gloire, G. Carrard, B. Friguet, Y. De Launoit, A. Burny, V. Bours, J. Piette, and C. Van Lint. 2003. Potentiation of tumor necrosis factor-induced NF-κB activation by deacetylase inhibitors is associated with a delayed cytoplasmic reappearance of IκBα. Mol. Cell. Biol. 23:6200-6209. - PMC - PubMed
-
- Aggarwal, B. B. 2003. Signalling pathways of the TNF superfamily: a double-edged sword. Nat. Rev. Immunol. 3:745-756. - PubMed
-
- Alterio de Goss, M., R. Holtappels, H.-P. Steffens, J. Podlech, P. Angele, L. Dreher, D. Thomas, and M. J. Reddehase. 1998. Control of cytomegalovirus in bone marrow transplantation chimeras lacking the prevailing antigen-presenting molecule in recipient tissues rests primarily on recipient-derived CD8 T cells. J. Virol. 72:7733-7744. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

