Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray
- PMID: 15596828
- PMCID: PMC538696
- DOI: 10.1128/JVI.79.1.341-352.2005
Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray
Abstract
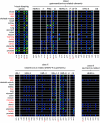
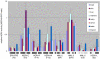
Retrovirus-like sequences account for 8 to 9% of the human genome. Among these sequences, about 8,000 pol-containing proviral elements have been identified to date. As part of our ongoing search for active and possibly disease-relevant human endogenous retroviruses (HERVs), we have recently developed an oligonucleotide-based microarray. The assay allows for both the detection and the identification of most known retroviral reverse transcriptase (RT)-related nucleic acids in biological samples. In the present study, we have investigated the transcriptional activity of representative members of 20 HERV families in 19 different normal human tissues. Qualitative evaluation of chip hybridization signals and quantitative analysis by real-time RT-PCR revealed distinct HERV activity in the human tissues under investigation, suggesting that HERV elements are active in human cells in a tissue-specific manner. Most active members of HERV families were found in mRNA prepared from skin, thyroid gland, placenta, and tissues of reproductive organs. In contrast, only few active HERVs were detectable in muscle cells. Human tissues that lack HERV transcription could not be found, confirming that human endogenous retroviruses are permanent components of the human transcriptome. Distinct activity patterns may reflect the characteristics of the regulatory machinery in these cells, e.g., cell type-dependent occurrence of transcriptional regulatory factors.
Figures
References
-
- Anderssen, S., E. Sjottem, G. Svineng, and T. Johansen. 1997. Comparative analyses of LTRs of the ERV-H family of primate-specific retrovirus-like elements isolated from marmoset, African green monkey, and man. Virology 234:14-30. - PubMed
-
- Andersson, A. C., P. J. Venables, R. R. Tonjes, J. Scherer, L. Eriksson, and E. Larsson. 2002. Developmental expression of HERV-R (ERV3) and HERV-K in human tissue. Virology 297:220-225. - PubMed
-
- Andersson, M. L., P. Medstrand, H. Yin, and J. Blomberg. 1996. Differential expression of human endogenous retroviral sequences similar to mouse mammary tumor virus in normal peripheral blood mononuclear cells. AIDS Res. Hum. Retrovir. 12:833-840. - PubMed
-
- Armbruester, V., M. Sauter, E. Krautkraemer, E. Meese, A. Kleiman, B. Best, K. Roemer, and N. Mueller-Lantzsch. 2002. A novel gene from the human endogenous retrovirus K expressed in transformed cells. Clin. Cancer Res. 8:1800-1807. - PubMed
-
- Barbulescu, M., G. Turner, M. I. Seaman, A. S. Deinard, K. K. Kidd, and J. Lenz. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9:861-868. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

