Inverted terminal repeat sequences are important for intermolecular recombination and circularization of adeno-associated virus genomes
- PMID: 15596830
- PMCID: PMC538689
- DOI: 10.1128/JVI.79.1.364-379.2005
Inverted terminal repeat sequences are important for intermolecular recombination and circularization of adeno-associated virus genomes
Abstract
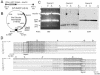
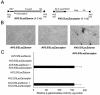
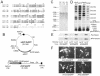
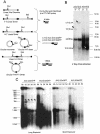
The relatively small package capacity (less than 5 kb) of adeno-associated virus (AAV) vectors has been effectively doubled with the development of dual-vector heterodimerization approaches. However, the efficiency of such dual-vector systems is limited not only by the extent to which intermolecular recombination occurs between two independent vector genomes, but also by the directional bias required for successful transgene reconstitution following concatemerization. In the present study, we sought to evaluate the mechanisms by which inverted terminal repeat (ITR) sequences mediate intermolecular recombination of AAV genomes, with the goal of engineering more efficient vectors for dual-vector trans-splicing approaches. To this end, we generated a novel AAV hybrid-ITR vector characterized by an AAV-2 and an AAV-5 ITR at opposite ends of the viral genome. This hybrid genome was efficiently packaged into either AAV-2 or AAV-5 capsids to generate infectious virions. Hybrid AV2:5 ITR viruses had a significantly lower capacity to form circular intermediates in infected cells than homologous AV2:2 and AV5:5 ITR vectors despite their similar capacity to express an encoded enhanced green fluorescent protein (EGFP) transgene. To examine whether the divergent ITR sequences contained within hybrid AV2:5 ITR vectors could direct intermolecular recombination in a tail-to-head fashion, we generated two hybrid ITR trans-splicing vectors (AV5:2LacZdonor and AV2:5LacZacceptor). Each delivered one exon of a beta-galactosidase minigene flanked by donor or acceptor splice sequences. These hybrid trans-splicing vectors were compared to homologous AV5:5 and AV2:2 trans-splicing vector sets for their ability to reconstitute beta-galactosidase gene expression. Results from this comparison demonstrated that hybrid ITR dual-vector sets had a significantly enhanced trans-splicing efficiency (6- to 10-fold, depending on the capsid serotype) compared to homologous ITR vectors. Molecular studies of viral genome structures suggest that hybrid ITR vectors provide more efficient directional recombination due to an increased abundance of linear-form genomes. These studies provide direct evidence for the importance of ITR sequences in directing intermolecular and intramolecular homologous recombination of AAV genomes. The use of hybrid ITR AAV vector genomes provides new strategies to manipulate viral genome conversion products and to direct intermolecular recombination events required for efficient dual-AAV vector reconstitution of the transgene.
Figures


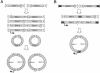
References
-
- Berns, K. I., and C. Giraud. 1996. Adeno-associated virus (AAV) vectors in gene therapy. Springer, New York, N.Y. - PubMed
-
- Carter, P. J., and R. J. Samulski. 2000. Adeno-associated viral vectors as gene delivery vehicles. Int. J. Mol. Med. 6:17-27. - PubMed
-
- Chao, H., Y. Liu, J. Rabinowitz, C. Li, R. J. Samulski, and C. E. Walsh. 2000. Several log increase in therapeutic transgene delivery by distinct adeno-associated viral serotype vectors. Mol. Ther. 2:619-623. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

