Evolutionary trace residues in noroviruses: importance in receptor binding, antigenicity, virion assembly, and strain diversity
- PMID: 15596848
- PMCID: PMC538680
- DOI: 10.1128/JVI.79.1.554-568.2005
Evolutionary trace residues in noroviruses: importance in receptor binding, antigenicity, virion assembly, and strain diversity
Abstract
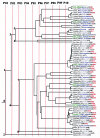
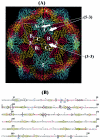
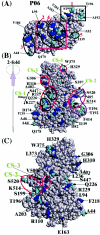
Noroviruses cause major epidemic gastroenteritis in humans. A large number of strains of these single-stranded RNA viruses have been reported. Due to the absence of infectious clones of noroviruses and the high sequence variability in their capsids, it has not been possible to identify functionally important residues in these capsids. Consequently, norovirus strain diversity is not understood on the basis of capsid functions, and the development of therapeutic compounds has been hampered. To determine functionally important residues in noroviruses, we have analyzed a number of norovirus capsid sequences in the context of the Norwalk virus capsid crystal structure by using the evolutionary trace method. This analysis has identified capsid protein residues that uniquely characterize different norovirus strains and provide new insights into capsid assembly and disassembly pathways and the strain diversity of these viruses. Such residues form specific three-dimensional clusters that may be of functional importance in noroviruses. One of these clusters includes residues known to participate in the proteolytic cleavage of these viruses at high pH. Other clusters are formed in capsid regions known to be important in the binding of antibodies to noroviruses, thereby indicating residues that may be important in the antigenicity of these viruses. The highly variable region of the capsid shows a distinct cluster whose residues may participate in norovirus-receptor interactions.
Figures


References
-
- Altschuh, D., A. M. Lesk, A. C. Bloomer, and A. Klug. 1987. Correlation of co-ordinated amino acid substitutions with function in viruses related to tobacco mosaic virus. J. Mol. Biol. 193:693-707. - PubMed
-
- Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of Norwalk-like viruses. J. Infect. Dis. 181(Suppl. 2):S336-S348. - PubMed
-
- Atchley, W. R., K. R. Wollenberg, W. M. Fitch, W. Terhalle, and A. W. Dress. 2000. Correlations among amino acid sites in bHLH protein domains: an information theoretic analysis. Mol. Biol. Evol. 17:164-178. - PubMed
-
- Blakeney, S. J., A. Cahill, and P. A. Reilly. 2003. Processing of Norwalk virus nonstructural proteins by a 3C-like cysteine proteinase. Virology 308:216-224. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

