A systematic characterization of mitochondrial proteome from human T leukemia cells
- PMID: 15598749
- PMCID: PMC1487188
- DOI: 10.1074/mcp.M400115-MCP200
A systematic characterization of mitochondrial proteome from human T leukemia cells
Abstract
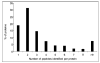
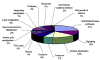
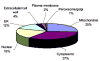
Global understanding of tissue-specific differences in mitochondrial signal transduction requires comprehensive mitochondrial protein identification from multiple cell and tissue types. Here, we explore the feasibility and efficiency of protein identification using the one-dimensional gel electrophoresis in combination with the nano liquid-chromatography tandem mass spectrometry (GeLC-MS/MS). The use of only 40 mug of purified mitochondrial proteins and data analysis using stringent scoring criteria and the molecular mass validation of the gel slices enables the identification of 227 known mitochondrial proteins (membrane and soluble) and 453 additional proteins likely to be associated with mitochondria. Replicate analyses of 60 mug of mitochondrial proteins on the faster scanning LTQ mass spectrometer validate all the previously identified proteins and most of the single hit proteins except the 81 single hit proteins. Among the identified proteins, 466 proteins are known to functionally participate in various processes such as respiration, tricarboxylic acid cycle (TCA cycle), amino acid and nucleotide metabolism, glycolysis, protection against oxidative stress, mitochondrial assembly, molecular transport, protein biosynthesis, cell cycle control, and many known cellular processes. The distribution of identified proteins in terms of size, pI, and hydrophobicity reveal that the present analytical strategy is largely unbiased and very efficient. Thus, we conclude that this approach is suitable for characterizing subcellular proteomes form multiple cells and tissues.
Figures


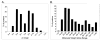

References
-
- McDonald TG, Van Eyk JE. Mitochondrial proteomics: Undercover in the lipid bilayer. Basic Res Cardiol. 2003;98:219–227. - PubMed
-
- Lopez MF, Melov S. Applied proteomics: Mitochondrial proteins and effect on functions. Circ Res. 2002;90:380–389. - PubMed
-
- Westermann B, Neupart W. “Omimics of the mitochondrion”: Two complementary proteomics approaches promise to move us closer to definition of the complete complement of proteins that make up a mitochondria”. Nat Biotechnol. 2003;21:239–240. - PubMed
-
- Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Willy S, Murphy AN, Gaucher SP, Capaldi RA, Gibson BW, Ghos SS. Characterization of the human heart mitochondrial proteome. Nat Biotechnol. 2003;21:281–286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P20 GM065764-010003/GM/NIGMS NIH HHS/United States
- S10 RR019436-018525/RR/NCRR NIH HHS/United States
- P01 HL070694-036740/HL/NHLBI NIH HHS/United States
- R01 HL067569-04/HL/NHLBI NIH HHS/United States
- P01 HL 70694/HL/NHLBI NIH HHS/United States
- P01 HL070694-019003/HL/NHLBI NIH HHS/United States
- S10 RR019436-018524/RR/NCRR NIH HHS/United States
- S10 RR019436-018522/RR/NCRR NIH HHS/United States
- R01 HL067569-03/HL/NHLBI NIH HHS/United States
- P01 HL 67569/HL/NHLBI NIH HHS/United States
- S10 RR019436-018523/RR/NCRR NIH HHS/United States
- P41 RR013186/RR/NCRR NIH HHS/United States
- P01 HL070694/HL/NHLBI NIH HHS/United States
- S10 RR019436-01/RR/NCRR NIH HHS/United States
- R01 HL067569/HL/NHLBI NIH HHS/United States
- RR 13186/RR/NCRR NIH HHS/United States
- P01 HL070694-010004/HL/NHLBI NIH HHS/United States
- P01 HL070694-036744/HL/NHLBI NIH HHS/United States
- R01 HL067569-05/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

