The plastid division proteins, FtsZ1 and FtsZ2, differ in their biochemical properties and sub-plastidial localization
- PMID: 15601251
- PMCID: PMC1134996
- DOI: 10.1042/BJ20041281
The plastid division proteins, FtsZ1 and FtsZ2, differ in their biochemical properties and sub-plastidial localization
Abstract
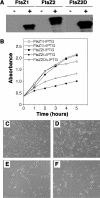
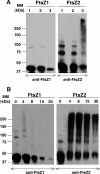
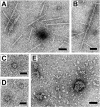
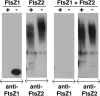
Plastid division in higher plants is morphologically similar to bacterial cell division, with a process termed binary fission involving constriction of the envelope membranes. FtsZ proteins involved in bacterial division are also present in higher plants, in which the ftsZ genes belong to two distinct families: ftsZ1 and ftsZ2. However, the roles of the corresponding proteins FtsZ1 and FtsZ2 in plastid division have not been determined. Here we show that the expression of plant FtsZ1 and FtsZ2 in bacteria has different effects on cell division, and that distinct protein domains are involved in the process. We have studied the assembly of purified FtsZ1 and FtsZ2 using a chemical cross-linking approach followed by PAGE and electron microscopy analyses of the resulting polymers. This has revealed that FtsZ1 is capable of forming long rod-shaped polymers and rings similar to the bacterial FtsZ structures, whereas FtsZ2 does not form any organized polymer. Moreover, using purified sub-plastidial fractions, we show that both proteins are present in the stroma, and that a subset of FtsZ2 is tightly bound to the purified envelope membranes. These results indicate that FtsZ2 has a localization pattern distinct from that of FtsZ1, which can be related to distinct properties of the proteins. From the results presented here, we propose a model for the sequential topological localization and functions of green plant FtsZ1 and FtsZ2 in chloroplast division.
Figures



Similar articles
-
In vivo quantitative relationship between plastid division proteins FtsZ1 and FtsZ2 and identification of ARC6 and ARC3 in a native FtsZ complex.Biochem J. 2008 Jun 1;412(2):367-78. doi: 10.1042/BJ20071354. Biochem J. 2008. PMID: 18284374
-
Colocalization of plastid division proteins in the chloroplast stromal compartment establishes a new functional relationship between FtsZ1 and FtsZ2 in higher plants.Plant Physiol. 2001 Dec;127(4):1656-66. Plant Physiol. 2001. PMID: 11743110 Free PMC article.
-
Arabidopsis FtsZ2-1 and FtsZ2-2 are functionally redundant, but FtsZ-based plastid division is not essential for chloroplast partitioning or plant growth and development.Mol Plant. 2009 Nov;2(6):1211-22. doi: 10.1093/mp/ssp077. Epub 2009 Sep 18. Mol Plant. 2009. PMID: 19995726
-
Plastid division: evolution, mechanism and complexity.Ann Bot. 2007 Apr;99(4):565-79. doi: 10.1093/aob/mcl249. Epub 2006 Nov 30. Ann Bot. 2007. PMID: 17138581 Free PMC article. Review.
-
The molecular biology of plastid division in higher plants.J Exp Bot. 2005 Apr;56(414):1061-77. doi: 10.1093/jxb/eri118. Epub 2005 Mar 7. J Exp Bot. 2005. PMID: 15753112 Review.
Cited by
-
Chloroplast division protein ARC3 regulates chloroplast FtsZ-ring assembly and positioning in arabidopsis through interaction with FtsZ2.Plant Cell. 2013 May;25(5):1787-802. doi: 10.1105/tpc.113.111047. Epub 2013 May 28. Plant Cell. 2013. PMID: 23715471 Free PMC article.
-
An emerging picture of plastid division in higher plants.Planta. 2005 Dec;223(1):1-4. doi: 10.1007/s00425-005-0078-y. Epub 2005 Sep 1. Planta. 2005. PMID: 16136332 No abstract available.
-
Two mechanosensitive channel homologs influence division ring placement in Arabidopsis chloroplasts.Plant Cell. 2011 Aug;23(8):2939-49. doi: 10.1105/tpc.111.088112. Epub 2011 Aug 2. Plant Cell. 2011. PMID: 21810996 Free PMC article.
-
Targeted gene knockouts reveal overlapping functions of the five Physcomitrella patens FtsZ isoforms in chloroplast division, chloroplast shaping, cell patterning, plant development, and gravity sensing.Mol Plant. 2009 Nov;2(6):1359-72. doi: 10.1093/mp/ssp076. Epub 2009 Sep 10. Mol Plant. 2009. PMID: 19946616 Free PMC article.
-
Plastid division control: the PDV proteins regulate DRP5B dynamin activity.Plant Mol Biol. 2013 Jun;82(3):255-66. doi: 10.1007/s11103-013-0059-7. Epub 2013 Apr 18. Plant Mol Biol. 2013. PMID: 23595201
References
-
- Osteryoung K. W., Vierling E. Conserved cell and organelle division. Nature (London) 1995;376:473–474. - PubMed
-
- Miyagishima S., Nishida K., Kuroiwa T. An evolutionary puzzle: chloroplast and mitochondrial division rings. Trends Plant Sci. 2003;8:432–438. - PubMed
-
- Gray M. W. Origin and evolution of organelle genomes. Curr. Opin. Genet. Dev. 1993;3:884–890. - PubMed
-
- Cavalier-Smith T. Membrane heredity and early chloroplast evolution. Trends Plant Sci. 2000;5:174–182. - PubMed
-
- Osteryoung K. W., Nunnari J. The division of endosymbiotic organelles. Science. 2003;302:1698–1704. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

