Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP
- PMID: 15601822
- PMCID: PMC535918
- DOI: 10.1101/gad.1255704
Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP
Abstract
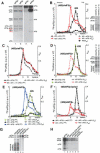
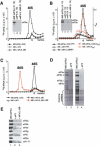
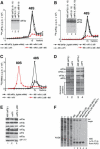
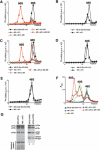
The 40S subunit in 48S complexes formed at the initiation codon of mRNA is bound to eukaryotic initiation factor (eIF) 3, eIF1, eIF1A, and an eIF2/GTP/Met-tRNAi(Met) ternary complex and can therefore not join a 60S subunit directly to form an 80S ribosome. We report that eIF5-induced hydrolysis of eIF2-bound GTP in 48S complexes led to release of eIF2-GDP but not eIF3 or eIF1. eIF5B did not influence factor release in the absence of 60S subunits. Therefore eIF3 and eIF1 dissociate from 40S subunits during, rather than before, the eIF5B-mediated subunit joining event. In the absence of eIF1, eIF5-stimulated hydrolysis of eIF2-bound GTP occurred at the same rate in 43S pre-initiation and 48S initiation complexes. GTP hydrolysis in 43S complexes assembled with eIF1 was much slower than in 43S or 48S complexes assembled without eIF1. Establishment of codon-anticodon base-pairing in 48S complexes relieved eIF1's inhibition. Thus, in addition to its role in initiation codon selection during 48S complex formation, eIF1 also participates in maintaining the fidelity of the initiation process at a later stage, hydrolysis of eIF2-bound GTP, by inhibiting premature GTP hydrolysis and by linking establishment of codon-anticodon base-pairing with GTP hydrolysis.
Figures


References
-
- Asano K.S., Phan, L., Anderson, J., and Hinnebusch, A.G. 1998. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from Saccharomyces cerevisiae. J. Biol. Chem. 273: 18573-18585. - PubMed
-
- ____. 1978. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J. Biol. Chem. 253: 3078-3087. - PubMed
-
- Carter A.P., Clemons, W.M., Brodersen, D.E., Morgan-Warren, R.J., Hartsch., T., Wimberly, B.T., and Ramakrishnan, V. 2001. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291: 498-501. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
