Translation of a small subset of Caenorhabditis elegans mRNAs is dependent on a specific eukaryotic translation initiation factor 4E isoform
- PMID: 15601834
- PMCID: PMC538781
- DOI: 10.1128/MCB.25.1.100-113.2005
Translation of a small subset of Caenorhabditis elegans mRNAs is dependent on a specific eukaryotic translation initiation factor 4E isoform
Abstract
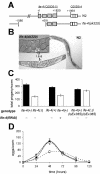
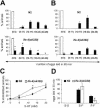
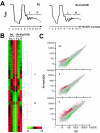
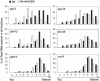
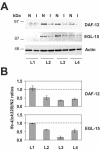
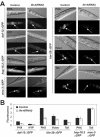
The mRNA cap-binding protein eukaryotic translation initiation factor 4E (eIF4E) participates in protein synthesis initiation, translational repression of specific mRNAs, and nucleocytoplasmic shuttling. Multiple isoforms of eIF4E are expressed in a variety of organisms, but their specific roles are poorly understood. We investigated one Caenorhabditis elegans isoform, IFE-4, which has homologues in plants and mammals. IFE-4::green fluorescent protein (GFP) was expressed in pharyngeal and tail neurons, body wall muscle, spermatheca, and vulva. Knockout of ife-4 by RNA interference (RNAi) or a null mutation produced a pleiotropic phenotype that included egg-laying defects. Sedimentation analysis demonstrated that IFE-4, but not IFE-1, was present in 48S initiation complexes, indicating that it participates in protein synthesis initiation. mRNAs affected by ife-4 knockout were determined by DNA microarray analysis of polysomal distribution. Polysome shifts, in the absence of total mRNA changes, were observed for only 33 of the 18,967 C. elegans mRNAs tested, of which a disproportionate number were related to egg laying and were expressed in neurons and/or muscle. Translational regulation was confirmed by reduced levels of DAF-12, EGL-15, and KIN-29. The functions of these proteins can explain some phenotypes observed in ife-4 knockout mutants. These results indicate that translation of a limited subset of mRNAs is dependent on a specific isoform of eIF4E.
Figures


References
-
- Bradley, C. A., J. C. Padovan, T. L. Thompson, C. A. Benoit, B. T. Chait, and R. E. Rhoads. 2002. Mass spectrometric analysis of the N-terminus of translational initiation factor eIF4G-1 reveals novel isoforms. J. Biol. Chem. 277:12559-12571. - PubMed
-
- Browning, K. S., C. Webster, J. K. M. Roberts, and J. M. Ravel. 1992. Identification of an isozyme form of protein synthesis initiation factor 4F in plants. J. Biol. Chem. 267:10096-10100. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
