The Saccharomyces cerevisiae Srb8-Srb11 complex functions with the SAGA complex during Gal4-activated transcription
- PMID: 15601835
- PMCID: PMC538787
- DOI: 10.1128/MCB.25.1.114-123.2005
The Saccharomyces cerevisiae Srb8-Srb11 complex functions with the SAGA complex during Gal4-activated transcription
Abstract
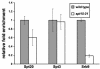
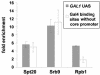
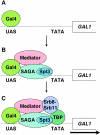
The Saccharomyces cerevisiae SAGA (Spt-Ada-Gcn5-acetyltransferase) complex functions as a coactivator during Gal4-activated transcription. A functional interaction between the SAGA component Spt3 and TATA-binding protein (TBP) is important for TBP binding at Gal4-activated promoters. To better understand the role of SAGA and other factors in Gal4-activated transcription, we selected for suppressors that bypass the requirement for SAGA. We obtained eight complementation groups and identified the genes corresponding to three of the groups as NHP10, HDA1, and SRB9. In contrast to the srb9 suppressor mutation that we identified, an srb9Delta mutation causes a strong defect in Gal4-activated transcription. Our studies have focused on this requirement for Srb9. Srb9 is part of the Srb8-Srb11 complex, associated with the Mediator coactivator. Srb8-Srb11 contains the Srb10 kinase, whose activity is important for GAL1 transcription. Our data suggest that Srb8-Srb11, including Srb10 kinase activity, is directly involved in Gal4 activation. By chromatin immunoprecipitation studies, Srb9 is present at the GAL1 promoter upon induction and facilitates the recruitment or stable association of TBP. Furthermore, the association of Srb9 with the GAL1 upstream activation sequence requires SAGA and specifically Spt3. Finally, Srb9 association also requires TBP. These results suggest that Srb8-Srb11 associates with the GAL1 promoter subsequent to SAGA binding, and that the binding of TBP and Srb8-Srb11 is interdependent.
Figures



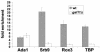

References
-
- Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1988. Current protocols in molecular biology. Greene Publishing Associates, New York, N.Y.
-
- Baganz, F., A. Hayes, D. Marren, D. C. Gardner, and S. G. Oliver. 1997. Suitability of replacement markers for functional analysis studies in Saccharomyces cerevisiae. Yeast 13:1563-1573. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
