BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase
- PMID: 15601839
- PMCID: PMC538799
- DOI: 10.1128/MCB.25.1.162-171.2005
BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase
Abstract
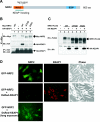
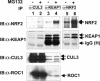
The concentrations and functions of many eukaryotic proteins are regulated by the ubiquitin pathway, which consists of ubiquitin activation (E1), conjugation (E2), and ligation (E3). Cullins are a family of evolutionarily conserved proteins that assemble by far the largest family of E3 ligase complexes. Cullins, via a conserved C-terminal domain, bind with the RING finger protein Roc1 to recruit the catalytic function of E2. Via a distinct N-terminal domain, individual cullins bind to a protein motif present in multiple proteins to recruit specific substrates. Cullin 3 (Cul3), but not other cullins, binds directly with BTB domains to constitute a potentially large number of BTB-CUL3-ROC1 E3 ubiquitin ligases. Here we report that the human BTB-Kelch protein Keap1, a negative regulator of the antioxidative transcription factor Nrf2, binds to CUL3 and Nrf2 via its BTB and Kelch domains, respectively. The KEAP1-CUL3-ROC1 complex promoted NRF2 ubiquitination in vitro and knocking down Keap1 or CUL3 by short interfering RNA resulted in NRF2 protein accumulation in vivo. We suggest that Keap1 negatively regulates Nrf2 function in part by targeting Nrf2 for ubiquitination by the CUL3-ROC1 ligase and subsequent degradation by the proteasome. Blocking NRF2 degradation in cells expressing both KEAP1 and NRF2 by either inhibiting the proteasome activity or knocking down Cul3, resulted in NRF2 accumulation in the cytoplasm. These results may reconcile previously observed cytoplasmic sequestration of NRF2 by KEAP1 and suggest a possible regulatory step between KEAP1-NRF2 binding and NRF2 degradation.
Figures





References
-
- Bai, C., P. Sen, K. Hofmann, L. Ma, M. Goebl, J. W. Harper, and S. J. Elledge. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86:263-274. - PubMed
-
- Bloom, D. A., and A. K. Jaiswal. 2003. Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H:quinone oxidoreductase-1 gene expression. J. Biol. Chem. 278:44675-44682. - PubMed
-
- Chen, A., K. Wu, S. Y. Fuchs, P. Tan, C. Gomez, and Z. Q. Pan. 2000. The conserved RING-H2 finger of ROC1 is required for ubiquitin ligation. J. Biol. Chem. 275:15432-15439. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
