Generation and characterization of endonuclease G null mice
- PMID: 15601850
- PMCID: PMC538798
- DOI: 10.1128/MCB.25.1.294-302.2005
Generation and characterization of endonuclease G null mice
Abstract
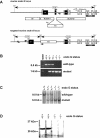
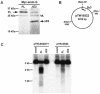
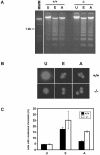
Endonuclease G (endo G) is one of the most abundant nucleases in eukaryotic cells. It is encoded in the nucleus and imported to the mitochondrial intermembrane space. This nuclease is active on single- and double-stranded DNA. We genetically disrupted the endo G gene in mice without disturbing a conserved, overlapping gene of unknown function that is oriented tail to tail with the endo G gene. In these mice, the production of endo G protein is not detected, and the disruption abolishes the nuclease activity of endo G. The absence of endo G has no effect on mitochondrial DNA copy number, structure, or mutation rate over the first five generations. There is also no obvious effect on nuclear DNA degradation in standard apoptosis assays. The endo G null mice are viable and show no age-related or generational abnormalities anatomically or histologically. We infer that this highly conserved protein has no mitochondrial or apoptosis function that can discerned by the assays described here and that it may have a function yet to be determined. The early embryonic lethality of endo G null mice recently reported by others may be due to the disruption of the gene that overlaps the endo G gene.
Figures

Similar articles
-
EndoG is dispensable in embryogenesis and apoptosis.Cell Death Differ. 2006 Jul;13(7):1147-55. doi: 10.1038/sj.cdd.4401787. Epub 2005 Oct 21. Cell Death Differ. 2006. PMID: 16239930
-
Primers for mitochondrial DNA replication generated by endonuclease G.Science. 1993 Aug 6;261(5122):765-9. doi: 10.1126/science.7688144. Science. 1993. PMID: 7688144
-
Characterization and expression of the mouse endonuclease G gene.DNA Cell Biol. 1997 Sep;16(9):1111-22. doi: 10.1089/dna.1997.16.1111. DNA Cell Biol. 1997. PMID: 9324313
-
Discovery, regulation, and action of the major apoptotic nucleases DFF40/CAD and endonuclease G.J Cell Biochem. 2005 Apr 15;94(6):1078-87. doi: 10.1002/jcb.20409. J Cell Biochem. 2005. PMID: 15723341 Review.
-
Analysis of nuclear degradation during lens cell differentiation.Cell Death Differ. 1998 Apr;5(4):251-61. doi: 10.1038/sj.cdd.4400351. Cell Death Differ. 1998. PMID: 10200471 Review.
Cited by
-
Endonuclease G preferentially cleaves 5-hydroxymethylcytosine-modified DNA creating a substrate for recombination.Nucleic Acids Res. 2014 Dec 1;42(21):13280-93. doi: 10.1093/nar/gku1032. Epub 2014 Oct 29. Nucleic Acids Res. 2014. PMID: 25355512 Free PMC article.
-
Endonuclease G does not play an obligatory role in poly(ADP-ribose) polymerase-dependent cell death after transient focal cerebral ischemia.Am J Physiol Regul Integr Comp Physiol. 2010 Jul;299(1):R215-21. doi: 10.1152/ajpregu.00747.2009. Epub 2010 Apr 28. Am J Physiol Regul Integr Comp Physiol. 2010. PMID: 20427721 Free PMC article.
-
Crystal structure of the EndoG/EndoGI complex: mechanism of EndoG inhibition.Nucleic Acids Res. 2009 Nov;37(21):7312-20. doi: 10.1093/nar/gkp770. Nucleic Acids Res. 2009. PMID: 19783821 Free PMC article.
-
Sensitivity of human prostate cancer cells to chemotherapeutic drugs depends on EndoG expression regulated by promoter methylation.Cancer Lett. 2008 Oct 18;270(1):132-43. doi: 10.1016/j.canlet.2008.04.053. Epub 2008 Jun 18. Cancer Lett. 2008. PMID: 18565644 Free PMC article.
-
Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos.Exp Neurol. 2009 Aug;218(2):193-202. doi: 10.1016/j.expneurol.2009.03.020. Epub 2009 Mar 28. Exp Neurol. 2009. PMID: 19332058 Free PMC article. Review.
References
-
- Adachi, N., and M. R. Lieber. 2002. Bidirectional gene organization: a common architectural motif of the human genome. Cell 109:807-809. - PubMed
-
- Bibb, M. J., R. A. van Etten, C. T. Wright, M. W. Walberg, and D. A. Clayton. 1981. Sequence and gene organization of mouse mitochondrial DNA. Cell 26:167-180. - PubMed
-
- Cerritelli, S. M., E. G. Frolova, C. Feng, A. Grinberg, P. E. Love, and R. J. Crouch. 2003. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol. Cell 11:807-815. - PubMed
-
- Clayton, D. A. 1991. Replication and transcription of vertebrate mitochondrial DNA. Annu. Rev. Cell Biol. 7:453-478. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
