Genome-wide prediction of stop codon readthrough during translation in the yeast Saccharomyces cerevisiae
- PMID: 15602002
- PMCID: PMC545446
- DOI: 10.1093/nar/gkh1004
Genome-wide prediction of stop codon readthrough during translation in the yeast Saccharomyces cerevisiae
Abstract
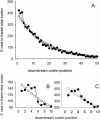
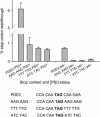
In-frame stop codons normally signal termination during mRNA translation, but they can be read as 'sense' (readthrough) depending on their context, comprising the 6 nt preceding and following the stop codon. To identify novel contexts directing readthrough, under-represented 5' and 3' stop codon contexts from Saccharomyces cerevisiae were identified by genome-wide survey in silico. In contrast with the nucleotide bias 3' of the stop codon, codon bias in the two codon positions 5' of the termination codon showed no correlation with known effects on stop codon readthrough. However, individually, poor 5' and 3' context elements were equally as effective in promoting stop codon readthrough in vivo, readthrough which in both cases responded identically to changes in release factor concentration. A novel method analysing specific nucleotide combinations in the 3' context region revealed positions +1,2,3,5 and +1,2,3,6 after the stop codon were most predictive of termination efficiency. Downstream of yeast open reading frames (ORFs), further in-frame stop codons were significantly over-represented at the +1, +2 and +3 codon positions after the ORF, acting to limit readthrough. Thus selection against stop codon readthrough is a dominant force acting on 3', but not on 5', nucleotides, with detectable selection on nucleotides as far downstream as +6 nucleotides. The approaches described can be employed to define potential readthrough contexts for any genome.
Figures



References
-
- Frolova L., Le Goff,X., Rasmussen,H.H., Cheperegin,S., Drugeon,G., Kress,M., Arman,I., Haenni,A.L., Celis,J.E., Philippe,M. et al. (1994) A highly conserved eukaryotic protein family possessing properties of polypeptide-chain release factor. Nature, 372, 701–703. - PubMed
-
- Stansfield I., Jones,K.M., Kushnirov,V.V., Dagkesamanskaya,A.R., Poznyakovski,A.I., Paushkin,S.V., Nierras,C.R., Cox,B.S., Teravanesyan,M.D. and Tuite,M.F. (1995) The products of the sup45 (erf1) and sup35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J., 14, 4365–4373. - PMC - PubMed
-
- Bertram G., Innes,S., Minella,O., Richardson,J. and Stansfield,I. (2001) Endless possibilities: translation termination and stop codon recognition. Microbiology, 147, 255–269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

