An extended transcriptional regulatory network of Escherichia coli and analysis of its hierarchical structure and network motifs
- PMID: 15604458
- PMCID: PMC545451
- DOI: 10.1093/nar/gkh1009
An extended transcriptional regulatory network of Escherichia coli and analysis of its hierarchical structure and network motifs
Abstract
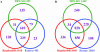
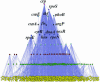
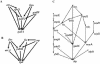
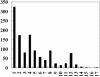
Recent studies of genome-wide transcriptional regulatory network (TRN) revealed several intriguing structural and dynamic features of gene expression at a system level. Unfortunately, the network under study is often far from complete. A critical question is thus how much the network is incomplete and to what extent this would affect the results of analysis. Here we compare the Escherichia coli TRN built by Shen-Orr et al. (Nature Genet., 31, 64-68) with two TRNs reconstructed from RegulonDB and Ecocyc respectively and present an extended E.coli TRN by integrating information from these databases and literature. The scale of the extended TRN is about twice as large as the previous ones. The new network preserves the multi-layer hierarchical structure which we recently reported but has more layers. More global regulators are inferred. While the feed forward loop (FFL) is confirmed to be highly representative in the network, the distribution of the different types of FFLs is different from that based on the incomplete network. In contrast to the notion of motif aggregation and formation of homologous motif clusters, we found that most FFLs interact and form a giant motif cluster. Furthermore, we show that only a small portion of the genes is solely regulated by only one FFL. Many genes are regulated by two or more interacting FFLs or other more complicated network motifs together with transcriptional factors not belonging to any network motifs, thereby forming complex regulatory circuits. Overall, the extended TRN represents a more solid basis for structural and functional analysis of genome-wide gene regulation in E.coli.
Figures
References
-
- Herrgard M.J. and Palsson,B.O. (2004) Flagellar biosynthesis in silico: building quantitative models of regulatory networks. Cell, 117, 689–690. - PubMed
-
- Luscombe N.M., Babu,M.M., Yu,H., Snyder,M., Teichmann,S.A. and Gerstein,M. (2004) Genomic analysis of regulatory network dynamics reveals large topological changes. Nature, 431, 308–312. - PubMed
-
- Milo R., Shen-Orr,S., Itzkovitz,S., Kashtan,N., Chklovskii,D. and Alon,U. (2002) Network motifs: simple building blocks of complex networks. Science, 298, 824–827. - PubMed
-
- Babu M.M., Luscombe,N.M., Aravind,L., Gerstein,M. and Teichmann,S.A. (2004) Structure and evolution of transcriptional regulatory networks. Curr. Opin. Struct. Biol., 14, 283–291. - PubMed
-
- Martinez-Antonio A. and Collado-Vides,J. (2003) Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol., 6, 482–489. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

