FunShift: a database of function shift analysis on protein subfamilies
- PMID: 15608176
- PMCID: PMC540021
- DOI: 10.1093/nar/gki067
FunShift: a database of function shift analysis on protein subfamilies
Abstract
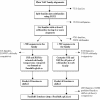
Members of a protein family normally have a general biochemical function in common, but frequently one or more subgroups have evolved a slightly different function, such as different substrate specificity. It is important to detect such function shifts for a more accurate functional annotation. The FunShift database described here is a compilation of function shift analysis performed between subfamilies in protein families. It consists of two main components: (i) subfamilies derived from protein domain families and (ii) pairwise subfamily comparisons analyzed for function shift. The present release, FunShift 12, was derived from Pfam 12 and consists of 151,934 subfamilies derived from 7300 families. We carried out function shift analysis by two complementary methods on families with up to 500 members. From a total of 179,210 subfamily pairs, 62,384 were predicted to be functionally shifted in 2881 families. Each subfamily pair is provided with a markup of probable functional specificity-determining sites. Tools for searching and exploring the data are provided to make this database a valuable resource for protein function annotation. Knowledge of these functionally important sites will be useful for experimental biologists performing functional mutation studies. FunShift is available at http://FunShift.cgb.ki.se.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

