Pseudomonas aeruginosa Genome Database and PseudoCAP: facilitating community-based, continually updated, genome annotation
- PMID: 15608211
- PMCID: PMC540001
- DOI: 10.1093/nar/gki047
Pseudomonas aeruginosa Genome Database and PseudoCAP: facilitating community-based, continually updated, genome annotation
Abstract
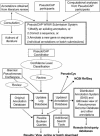
Using the Pseudomonas aeruginosa Genome Project as a test case, we have developed a database and submission system to facilitate a community-based approach to continually updated genome annotation (http://www.pseudomonas.com). Researchers submit proposed annotation updates through one of three web-based form options which are then subjected to review, and if accepted, entered into both the database and log file of updates with author acknowledgement. In addition, a coordinator continually reviews literature for suitable updates, as we have found such reviews to be the most efficient. Both the annotations database and updates-log database have Boolean search capability with the ability to sort results and download all data or search results as tab-delimited files. To complement this peer-reviewed genome annotation, we also provide a linked GBrowse view which displays alternate annotations. Additional tools and analyses are also integrated, including PseudoCyc, and knockout mutant information. We propose that this database system, with its focus on facilitating flexible queries of the data and providing access to both peer-reviewed annotations as well as alternate annotation information, may be a suitable model for other genome projects wishing to use a continually updated, community-based annotation approach. The source code is freely available under GNU General Public Licence.
Figures
References
-
- Stover C.K., Pham,X.Q., Erwin,A.L., Mizoguchi,S.D., Warrener,P., Hickey,M.J., Brinkman,F.S., Hufnagle,W.O., Kowalik,D.J., Lagrou,M., Garber,R.L., Goltry,L., Tolentino,E., Westbrock-Wadman,S., Yuan,Y., Brody,L.L., Coulter,S.N., Folger,K.R., Kas,A., Larbig,K., Lim,R., Smith,K., Spencer,D., Wong,G.K., Wu,Z., Paulsen,I.T., Reizer,J., Saier,M.H., Hancock,R.E., Lory,S. and Olson,M.V. (2000) Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature, 406, 959–964. - PubMed
-
- Brinkman F.S., Hancock,R.E. and Stover,C.K. (2000) Sequencing solution: use volunteer annotators organized via Internet. Nature, 406, 933. - PubMed
-
- Williams N. (1995) Closing in on the complete yeast genome sequence. Science, 268, 1560–1561. - PubMed
-
- Williams N. (1996) Yeast genome sequence ferments new research. Science, 272, 481. - PubMed
-
- Goffeau A., Barrell,B.G., Bussey,H., Davis,R.W., Dujon,B., Feldmann,H., Galibert,F., Hoheisel,J.D., Jacq,C., Johnston,M. et al. (1996) Life with 6000 genes. Science, 274, 563–567. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases