HOPPSIGEN: a database of human and mouse processed pseudogenes
- PMID: 15608268
- PMCID: PMC540038
- DOI: 10.1093/nar/gki084
HOPPSIGEN: a database of human and mouse processed pseudogenes
Erratum in
- Nucleic Acids Res. 2005;33(1):448. Adel, Khelifi [corrected to Khelifi, Adel]; Laurent, Duret [corrected to Durent, Laurent]; Dominique, Mouchiroud [corrected to Mouchiroud, Dominique]
Abstract
Processed pseudogenes result from reverse transcribed mRNAs. In general, because processed pseudogenes lack promoters, they are no longer functional from the moment they are inserted into the genome. Subsequently, they freely accumulate substitutions, insertions and deletions. Moreover, the ancestral structure of processed pseudogenes could be easily inferred using the sequence of their functional homologous genes. Owing to these characteristics, processed pseudogenes represent good neutral markers for studying genome evolution. Recently, there is an increasing interest for these markers, particularly to help gene prediction in the field of genome annotation, functional genomics and genome evolution analysis (patterns of substitution). For these reasons, we have developed a method to annotate processed pseudogenes in complete genomes. To make them useful to different fields of research, we stored them in a nucleic acid database after having annotated them. In this work, we screened both mouse and human complete genomes from ENSEMBL to find processed pseudogenes generated from functional genes with introns. We used a conservative method to detect processed pseudogenes in order to minimize the rate of false positive sequences. Within processed pseudogenes, some are still having a conserved open reading frame and some have overlapping gene locations. We designated as retroelements all reverse transcribed sequences and more strictly, we designated as processed pseudogenes, all retroelements not falling in the two former categories (having a conserved open reading or overlapping gene locations). We annotated 5823 retroelements (5206 processed pseudogenes) in the human genome and 3934 (3428 processed pseudogenes) in the mouse genome. Compared to previous estimations, the total number of processed pseudogenes was underestimated but the aim of this procedure was to generate a high-quality dataset. To facilitate the use of processed pseudogenes in studying genome structure and evolution, DNA sequences from processed pseudogenes, and their functional reverse transcribed homologs, are now stored in a nucleic acid database, HOPPSIGEN. HOPPSIGEN can be browsed on the PBIL (Pole Bioinformatique Lyonnais) World Wide Web server (http://pbil.univ-lyon1.fr/) or fully downloaded for local installation.
Figures
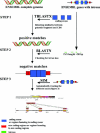


← If (SIM score > 20 and PR similar to at least two exons) ↠ PR is retained;
else if (SIM score > 20 and PR similar to one exon) ↠ PR is a single exon (case C);
else if (SIM score < 20) ↠ PR is discarded;
↑ for each splicing site: test if ai or ai±1 is near the splicing site Sj on the gene (Sj ± 10 bp);
If test succeeded: then if d(ai, ai+1) > 50 bp ↠ PR is an unprocessed paralog (case A and B);
if test failed ↠ PR is a retroelement (case D);


References
-
- Vanin E.F. (1985) Processed pseudogenes: characteristics and evolution. Annu. Rev. Genet., 19, 253–272. - PubMed
-
- Esnault C., Maestre,J. and Heidmann,T. (2000) Human LINE retrotransposons generate processed pseudogenes. Nature Genet., 24, 363–367. - PubMed
-
- Mighell A.J., Smith,N.R., Robinson,P.A. and Markham,A.F. (2000) Vertebrate pseudogenes. FEBS Lett., 468, 109–114. - PubMed
-
- Pavlicek A., Paces,J., Zika,R. and Hejnar,J. (2002) Length distribution of long interspersed nucleotide elements (LINEs) and processed pseudogenes of human endogenous retroviruses: implications for retrotransposition and pseudogene detection. Gene, 300, 189–194. - PubMed
-
- Crollius H.R., Jaillon,O., Dasilva,C., Ozouf-Costaz,C., Fizames,C., Fischer,C., Bouneau,L., Billault,A., Quetier,F., Saurin,W., Bernot,A. and Weissenbach,J. (2000) Characterization and repeat analysis of the compact genome of the freshwater pufferfish Tetraodon nigroviridis. Genome Res., 10, 939–949. - PMC - PubMed

