A NAC domain protein interacts with tomato leaf curl virus replication accessory protein and enhances viral replication
- PMID: 15608335
- PMCID: PMC544507
- DOI: 10.1105/tpc.104.027235
A NAC domain protein interacts with tomato leaf curl virus replication accessory protein and enhances viral replication
Abstract
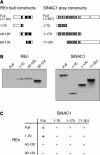
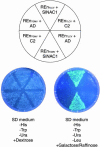
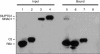
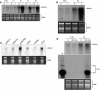
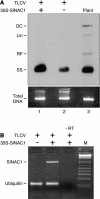
Geminivirus replication enhancer (REn) proteins dramatically increase the accumulation of viral DNA species by an unknown mechanism. In this study, we present evidence implicating SlNAC1, a new member of the NAC domain protein family from tomato (Solanum lycopersicum), in Tomato leaf curl virus (TLCV) REn function. We isolated SlNAC1 using yeast (Saccharomyces cerevisiae) two-hybrid technology and TLCV REn as bait, and confirmed the interaction between these proteins in vitro. TLCV induces SlNAC1 expression specifically in infected cells, and this upregulation requires REn. In a transient TLCV replication system, overexpression of SlNAC1 resulted in a substantial increase in viral DNA accumulation. SlNAC1 colocalized with REn to the nucleus and activated transcription of a reporter gene in yeast, suggesting that in healthy cells it functions as a transcription factor. Together, these results imply that SlNAC1 plays an important role in the process by which REn enhances TLCV replication.
Figures




References
-
- Abouzid, A.M., Frischmuth, T., and Jeske, H. (1988). A putative replicative form of the Abutilon mosaic virus (gemini group) in a chromatin-like structure. Mol. Gen. Genet. 212, 252–258.
-
- Boisnard-Lorig, C., Colon-Carmona, A., Bauch, M., Hodge, S., Doerner, P., Bancharel, E., Dumas, C., Haseloff, J., and Berger, F. (2001). Dynamic analyses of the expression of the HISTONE∷YFP fusion protein in arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13, 495–509. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

