How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures
- PMID: 15608617
- PMCID: PMC1299224
- DOI: 10.1038/sj.embor.7400314
How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures
Erratum in
- EMBO Rep. 2005 Feb;6(2):191
Abstract
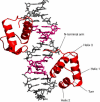
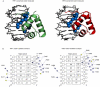
Human telomeres consist of tandem arrays of TTAGGG sequence repeats that are specifically bound by two proteins, TRF1 and TRF2. They bind to DNA as preformed homodimers and have the same architecture in which the DNA-binding domains (Dbds) form independent structural units. Despite these similarities, TRF1 and TRF2 have different functions at telomeres. The X-ray crystal structures of both TRF1- and TRF2-Dbds in complex with telomeric DNA (2.0 and 1.8 angstroms resolution, respectively) show that they recognize the same TAGGGTT binding site by means of homeodomains, as does the yeast telomeric protein Rap1p. Two of the three G-C base pairs that characterize telomeric repeats are recognized specifically and an unusually large number of water molecules mediate protein-DNA interactions. The binding of the TRF2-Dbd to the DNA double helix shows no distortions that would account for the promotion of t-loops in which TRF2 has been implicated.
Figures


References
-
- Abrahams JP, Leslie AGW (1996) Methods used in the structure determination of bovine mitochondrial F-1 ATPase. Acta Crystallogr D 52: 30–42 - PubMed
-
- Ancelin K, Brun C, Gilson E (1998) Role of the telomeric DNA-binding protein TRF2 in the stability of human chromosome ends. BioEssays 20: 879–883 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

