GRP1 pleckstrin homology domain: activation parameters and novel search mechanism for rare target lipid
- PMID: 15610010
- PMCID: PMC3625374
- DOI: 10.1021/bi049017a
GRP1 pleckstrin homology domain: activation parameters and novel search mechanism for rare target lipid
Abstract
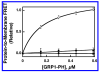
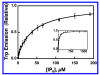
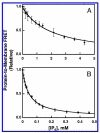
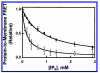
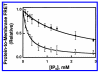
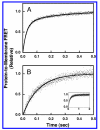
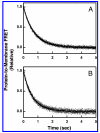
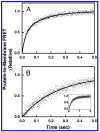
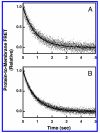
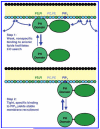
Pleckstrin homology (PH) domains play a central role in a wide array of signaling pathways by binding second messenger lipids of the phosphatidylinositol phosphate (PIP) lipid family. A given type of PIP lipid is formed in a specific cellular membrane where it is generally a minor component of the bulk lipid mixture. For example, the signaling lipid PI(3,4,5)P(3) (or PIP(3)) is generated primarily in the inner leaflet of the plasma membrane where it is believed to never exceed 0.02% of the bulk lipid. The present study focuses on the PH domain of the general receptor for phosphoinositides, isoform 1 (GRP1), which regulates the actin cytoskeleton in response to PIP(3) signals at the plasma membrane surface. The study systematically analyzes both the equilibrium and kinetic features of GRP1-PH domain binding to its PIP lipid target on a bilayer surface. Equilibrium binding measurements utilizing protein-to-membrane fluorescence resonance energy transfer (FRET) to detect GRP1-PH domain docking to membrane-bound PIP lipids confirm specific binding to PIP(3). A novel FRET competitive binding measurement developed to quantitate docking affinity yields a K(D) of 50 +/- 10 nM for GRP1-PH domain binding to membrane-bound PIP(3) in a physiological lipid mixture approximating the composition of the plasma membrane inner leaflet. This observed K(D) lies in a suitable range for regulation by physiological PIP(3) signals. Interestingly, the affinity of the interaction decreases at least 12-fold when the background anionic lipids phosphatidylserine (PS) and phosphatidylinositol (PI) are removed from the lipid mixture. Stopped-flow kinetic studies using protein-to-membrane FRET to monitor association and dissociation time courses reveal that this affinity decrease arises from a corresponding decrease in the on-rate for GRP1-PH domain docking with little or no change in the off-rate for domain dissociation from membrane-bound PIP(3). Overall, these findings indicate that the PH domain interacts not only with its target lipid, but also with other features of the membrane surface. The results are consistent with a previously undescribed type of two-step search mechanism for lipid binding domains in which weak, nonspecific electrostatic interactions between the PH domain and background anionic lipids facilitate searching of the membrane surface for PIP(3) headgroups, thereby speeding the high-affinity, specific docking of the domain to its rare target lipid.
Figures
Similar articles
-
Molecular mechanism of membrane binding of the GRP1 PH domain.J Mol Biol. 2013 Sep 9;425(17):3073-90. doi: 10.1016/j.jmb.2013.05.026. Epub 2013 Jun 6. J Mol Biol. 2013. PMID: 23747485 Free PMC article.
-
Membrane docking geometry of GRP1 PH domain bound to a target lipid bilayer: an EPR site-directed spin-labeling and relaxation study.PLoS One. 2012;7(3):e33640. doi: 10.1371/journal.pone.0033640. Epub 2012 Mar 30. PLoS One. 2012. PMID: 22479423 Free PMC article.
-
Finding a needle in a haystack: the role of electrostatics in target lipid recognition by PH domains.PLoS Comput Biol. 2012;8(7):e1002617. doi: 10.1371/journal.pcbi.1002617. Epub 2012 Jul 26. PLoS Comput Biol. 2012. PMID: 22844242 Free PMC article.
-
Cellular and molecular interactions of phosphoinositides and peripheral proteins.Chem Phys Lipids. 2014 Sep;182:3-18. doi: 10.1016/j.chemphyslip.2014.02.002. Epub 2014 Feb 17. Chem Phys Lipids. 2014. PMID: 24556335 Free PMC article. Review.
-
Regulation of V-ATPase Activity and Organelle pH by Phosphatidylinositol Phosphate Lipids.Front Cell Dev Biol. 2020 Jun 23;8:510. doi: 10.3389/fcell.2020.00510. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32656214 Free PMC article. Review.
Cited by
-
Microscopic Characterization of GRP1 PH Domain Interaction with Anionic Membranes.J Comput Chem. 2020 Mar 5;41(6):489-499. doi: 10.1002/jcc.26109. Epub 2019 Nov 25. J Comput Chem. 2020. PMID: 31762060 Free PMC article.
-
Phosphatidylinositol 4,5-bisphosphate-specific AKT1 is oncogenic.Int J Cancer. 2010 Jul 1;127(1):239-44. doi: 10.1002/ijc.25012. Int J Cancer. 2010. PMID: 19876913 Free PMC article.
-
Multiple lipid binding sites determine the affinity of PH domains for phosphoinositide-containing membranes.Sci Adv. 2020 Feb 19;6(8):eaay5736. doi: 10.1126/sciadv.aay5736. eCollection 2020 Feb. Sci Adv. 2020. PMID: 32128410 Free PMC article.
-
Biophysical methods for the characterization of PTEN/lipid bilayer interactions.Methods. 2015 May;77-78:125-35. doi: 10.1016/j.ymeth.2015.02.004. Epub 2015 Feb 16. Methods. 2015. PMID: 25697761 Free PMC article. Review.
-
A Complex Interplay of Anionic Phospholipid Binding Regulates 3'-Phosphoinositide-Dependent-Kinase-1 Homodimer Activation.Sci Rep. 2019 Oct 10;9(1):14527. doi: 10.1038/s41598-019-50742-8. Sci Rep. 2019. PMID: 31601855 Free PMC article.
References
-
- Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell. 2004;13:677–88. - PubMed
-
- Czech MP. Dynamics of phosphoinositides in membrane retrieval and insertion. Annu. ReV. Physiol. 2003;65:791–815. - PubMed
-
- Luo J, Manning BD, Cantley LC. Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer Cell. 2003;4:257–62. - PubMed
-
- Hurley JH, Meyer T. Subcellular targeting by membrane lipids. Curr. Opin. Cell Biol. 2001;13:146–52. - PubMed
-
- Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD. Synthesis and function of 3-phosphorylated inositol lipids. Annu. ReV. Biochem. 2001;70:535–602. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

