Misfolded BiP is degraded by a proteasome-independent endoplasmic-reticulum-associated degradation pathway
- PMID: 15610068
- PMCID: PMC1135023
- DOI: 10.1042/BJ20041312
Misfolded BiP is degraded by a proteasome-independent endoplasmic-reticulum-associated degradation pathway
Abstract
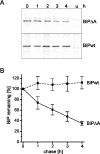
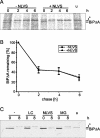
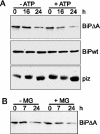
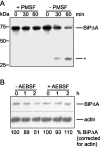
Misfolded proteins are removed from the ER (endoplasmic reticulum) by retrotranslocation to the cytosol and degradation by the ubiquitin-proteasome system in a process designated ERAD (ER-associated degradation). Analysing the turnover of a misfolded form of the ER-resident chaperone BiP (heavy-chain binding protein) (BiPDeltaA), we found that the degradation of BiPDeltaA did not follow this general ERAD pathway. In transfected cells, BiPDeltaA was degraded, although proteasome-dependent ERAD was inactivated either by proteasome inhibitors or by ATP depletion. In semi-permeabilized cells, which did not support the degradation of the proteasomal substrate alpha1-antitrypsin, the degradation of BiPDeltaA was still functional, excluding the Golgi apparatus or lysosomes as the degradative compartment. The degradation of BiPDeltaA was recapitulated in biosynthetically loaded brain microsomes and in an extract of luminal ER proteins. In contrast with proteasome-dependent ERAD, degradation fragments were detectable inside the microsomes and in the extract, and the degradation was prevented by a serine protease inhibitor. These results show that the degradation of BiPDeltaA was initiated in the ER lumen by a serine protease, and support the view that proteasome-independent ERAD pathways exist.
Figures




References
-
- Ellgaard L., Helenius A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003;4:181–191. - PubMed
-
- Trombetta E. S., Parodi A. J. Quality control and protein folding in the secretory pathway. Annu. Rev. Cell Dev. Biol. 2003;19:649–676. - PubMed
-
- Sitia R., Braakman I. Quality control in the endoplasmic reticulum protein factory. Nature (London) 2003;426:891–894. - PubMed
-
- Tsai B., Ye Y., Rapoport T. A. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell Biol. 2002;3:246–255. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

