AtNAP1 represents an atypical SufB protein in Arabidopsis plastids
- PMID: 15611066
- PMCID: PMC1401503
- DOI: 10.1074/jbc.M413082200
AtNAP1 represents an atypical SufB protein in Arabidopsis plastids
Abstract
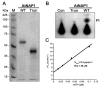
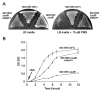
The assembly of iron-sulfur (Fe-S) clusters involves several pathways and in prokaryotes the mobilization of the sulfur (SUF) system is paramount for Fe-S biogenesis and repair during oxidative stress. The prokaryotic SUF system consists of six proteins: SufC is an ABC/ATPase that forms a complex with SufB and SufD, SufA acts as a scaffold protein, and SufE and SufS are involved in sulfur mobilization from cysteine. Despite the importance of Fe-S proteins in higher plant plastids, little is known regarding plastidic Fe-S cluster assembly. We have recently shown that Arabidopsis harbors an evolutionary conserved plastidic SufC protein (AtNAP7) capable of hydrolyzing ATP and interacting with the SufD homolog AtNAP6. Based on this and the prokaryotic SUF system we speculated that a SufB-like protein may exist in plastids. Here we demonstrate that the Arabidopsis plastid-localized SufB homolog AtNAP1 can complement SufB deficiency in Escherichia coli during oxidative stress. Furthermore, we demonstrate that AtNAP1 can interact with AtNAP7 inside living chloroplasts suggesting the presence of a plastidic AtNAP1.AtNAP6.AtNAP7 complex and remarkable evolutionary conservation of the SUF system. However, in contrast to prokaryotic SufB proteins with no associated ATPase activity we show that AtNAP1 is an iron-stimulated ATPase and that AtNAP1 is capable of forming homodimers. Our results suggest that AtNAP1 represents an atypical plastidic SufB-like protein important for Fe-S cluster assembly and for regulating iron homeostasis in Arabidopsis.
Figures



References
-
- Beinert H, Holm RH, Munck E. Science. 1997;277:653–659. - PubMed
-
- Beinert H, Kiley PJ. Curr Opin Chem Biol. 1999;3:152–157. - PubMed
-
- Lill R, Kispal G. Trends Biochem Sci. 2000;25:352–356. - PubMed
-
- Frazzon J, Dean DR. Curr Opin Chem Biol. 2003;7:166–173. - PubMed
-
- Takahashi Y, Tokumoto U. J Biol Chem. 2002;277:28380–28383. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

