Epstein-Barr virus gp42 is posttranslationally modified to produce soluble gp42 that mediates HLA class II immune evasion
- PMID: 15613312
- PMCID: PMC538541
- DOI: 10.1128/JVI.79.2.841-852.2005
Epstein-Barr virus gp42 is posttranslationally modified to produce soluble gp42 that mediates HLA class II immune evasion
Abstract
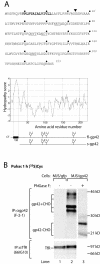
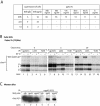
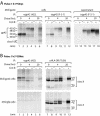
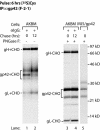
Epstein-Barr virus (EBV) resides as a persistent infection in human leukocyte antigen (HLA) class II+ B lymphocytes and is associated with a number of malignancies. The EBV lytic-phase protein gp42 serves at least two functions: gp42 acts as the coreceptor for viral entry into B cells and hampers T-cell recognition via HLA class II molecules through steric hindrance of T-cell receptor-class II-peptide interactions. Here, we show that gp42 associates with class II molecules at their various stages of maturation, including immature alphabetaIi heterotrimers and mature alphabeta-peptide complexes. When analyzing the biosynthesis and maturation of gp42 in cells stably expressing the viral protein, we found that gp42 occurs in two forms: a full-length type II membrane protein and a truncated soluble form. Soluble gp42 is generated by proteolytic cleavage in the endoplasmic reticulum and is secreted. Soluble gp42 is sufficient to inhibit HLA class II-restricted antigen presentation to T cells. In an almost pure population of Burkitt's lymphoma cells in the EBV lytic cycle, both transmembrane and soluble forms of gp42 are detected. These results imply that soluble gp42 is generated during EBV lytic infection and could contribute to undetected virus production by mediating evasion from T-cell immunity.
Figures



References
-
- Borza, C. M., and L. M. Hutt-Fletcher. 2002. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat. Med. 8:594-599. - PubMed
-
- Campadelli-Fiume, G., F. Cocchi, L. Menotti, and M. Lopez. 2000. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev. Med. Virol. 10:305-319. - PubMed
-
- Clark, A. E., and T. Yakoshi. 1984. HB10A, p. 195. In A. Bernard, L. Baunsell, J. Dausset, and S. Schlossman (ed.), Leukocyte typing. Springer-Verlag KG, Heidelberg, Germany.
-
- Cresswell, P. 1994. Assembly, transport, and function of MHC class II molecules. Annu. Rev. Immunol. 12:259-293. - PubMed
-
- Dalbey, R. E., and G. von Heijne. 1992. Signal peptidases in prokaryotes and eukaryotes—a new protease family. Trends Biochem. Sci. 17:474-478. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

