Efficient rescue of hepatitis C virus RNA replication by trans-complementation with nonstructural protein 5A
- PMID: 15613318
- PMCID: PMC538567
- DOI: 10.1128/JVI.79.2.896-909.2005
Efficient rescue of hepatitis C virus RNA replication by trans-complementation with nonstructural protein 5A
Abstract
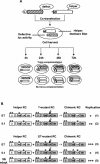
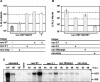
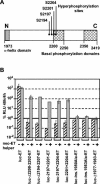
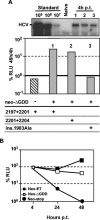
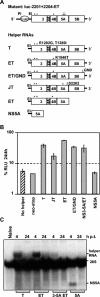
Studies of Hepatitis C virus (HCV) RNA replication have become possible with the development of subgenomic replicons. This system allows the functional analysis of the essential components of the viral replication complex, which so far are poorly defined. In the present study we wanted to investigate whether lethal mutations in HCV nonstructural genes can be rescued by trans-complementation. Therefore, a series of replicon RNAs carrying mutations in NS3, NS4B, NS5A, and NS5B that abolish replication were transfected into Huh-7 hepatoma cells harboring autonomously replicating helper RNAs. Similar to data described for the Bovine viral diarrhea virus (C. W. Grassmann, O. Isken, N. Tautz, and S. E. Behrens, J. Virol. 75:7791-7802, 2001), we found that only NS5A mutants could be efficiently rescued. There was no evidence for RNA recombination between helper and mutant RNAs, and we did not observe reversions in the transfected mutants. Furthermore, we established a transient complementation assay based on the cotransfection of helper and mutant RNAs. Using this assay, we extended our results and demonstrated that (i) inactivating NS5A mutations affecting the amino-terminal amphipathic helix cannot be complemented in trans; (ii) replication of the helper RNA is not necessary to allow efficient trans-complementation; and (iii) the minimal sequence required for trans-complementation of lethal NS5A mutations is NS3 to -5A, whereas NS5A expressed alone does not restore RNA replication. In summary, our results provide the first insight into the functional organization of the HCV replication complex.
Figures



References
-
- Arima, N., C. Y. Kao, T. Licht, R. Padmanabhan, Y. Sasaguri, and R. Padmanabhan. 2001. Modulation of cell growth by the hepatitis C virus nonstructural protein NS5A. J Biol. Chem. 276:12675-12684. - PubMed
-
- Bartenschlager, R., M. Frese, and T. Pietschmann. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63:71-180. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

