Studying multiprotein complexes by multisignal sedimentation velocity analytical ultracentrifugation
- PMID: 15613487
- PMCID: PMC538923
- DOI: 10.1073/pnas.0408399102
Studying multiprotein complexes by multisignal sedimentation velocity analytical ultracentrifugation
Abstract
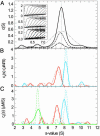
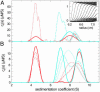
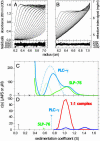
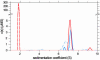
Protein interactions can promote the reversible assembly of multiprotein complexes, which have been identified as critical elements in many regulatory processes in cells. The biophysical characterization of assembly products, their number and stoichiometry, and the dynamics of their interactions in solution can be very difficult. A classical first-principle approach for the study of purified proteins and their interactions is sedimentation velocity analytical ultracentrifugation. This approach allows one to distinguish different protein complexes based on their migration in the centrifugal field without isolating reversibly formed complexes from the individual components. An important existing limitation for systems with multiple components and assembly products is the identification of the species associated with the observed sedimentation rates. We developed a computational approach for integrating multiple optical signals into the sedimentation coefficient distribution analysis of components, which combines the size-dependent hydrodynamic separation with discrimination of the extinction properties of the sedimenting species. This approach allows one to deduce the stoichiometry and to assign the identity of the assembly products without prior assumptions of the number of species and the nature of their interaction. Although chromophoric labels may be used to enhance the spectral resolution, we demonstrate the ability to work label-free for three-component protein mixtures. We observed that the spectral discrimination can synergistically enhance the hydrodynamic resolution. This method can take advantage of differences in the absorbance spectra of interacting solution components, for example, for the study of protein-protein, protein-nucleic acid or protein-small molecule interactions, and can determine the size, hydrodynamic shape, and stoichiometry of multiple complexes in solution.
Figures
References
-
- Heller, M., Goodlett, D. R., Watts, J. D. & Aebersold, R. (2000) Electrophoresis 21, 2180-2195. - PubMed
-
- Herr, A. B., White, C. L., Milburn, C., Wu, C. & Bjorkman, P. J. (2003) J. Mol. Biol. 327, 645-657. - PubMed
-
- Davis, M. M., Krogsgaard, M., Huppa, J. B., Sumen, C., Purbhoo, M. A., Irvine, D. J., Wu, L. C. & Ehrlich, L. (2003) Annu. Rev. Biochem. 72, 717-742. - PubMed
-
- Andersen, P. S., Schuck, P., Sundberg, E. J., Geisler, C., Karjalainen, K. & Mariuzza, R. A. (2002) Biochemistry 41, 5177-5184. - PubMed
-
- Bers, D. M. (2004) J. Mol. Cell Cardiol. 37, 417-429. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

