GERBIL: Genotype resolution and block identification using likelihood
- PMID: 15615859
- PMCID: PMC544046
- DOI: 10.1073/pnas.0404730102
GERBIL: Genotype resolution and block identification using likelihood
Abstract
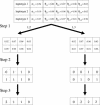
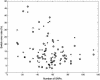
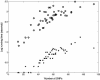
The abundance of genotype data generated by individual and international efforts carries the promise of revolutionizing disease studies and the association of phenotypes with individual polymorphisms. A key challenge is providing an accurate resolution (phasing) of the genotypes into haplotypes. We present here results on a method for genotype phasing in the presence of recombination. Our analysis is based on a stochastic model for recombination-poor regions ("blocks"), in which haplotypes are generated from a small number of core haplotypes, allowing for mutations, rare recombinations, and errors. We formulate genotype resolution and block partitioning as a maximum-likelihood problem and solve it by an expectation-maximization algorithm. The algorithm was implemented in a software package called GERBIL (genotype resolution and block identification using likelihood), which is efficient and simple to use. We tested GERBIL on four large-scale sets of genotypes. It outperformed two state-of-the-art phasing algorithms. The phase algorithm was slightly more accurate than GERBIL when allowed to run with default parameters, but required two orders of magnitude more time. When using comparable running times, GERBIL was consistently more accurate. For data sets with hundreds of genotypes, the time required by phase becomes prohibitive. We conclude that GERBIL has a clear advantage for studies that include many hundreds of genotypes and, in particular, for large-scale disease studies.
Figures
References
-
- Patil, N., Berno, A. J., Hinds, D. A., Barrett, W. A., Doshi, J. M., Hacker, C. R., Kautzer, C. R., Lee, D. H., Marjoribanks, C., McDonough, D. P., et al. (2001) Science 294, 1719-1723. - PubMed
-
- Gabriel, S. B., Schaffner, S. F., Nguyen, H., Moore, J. M., Roy, J., Blumenstiel, B., Higgins, J., DeFelice, M., Lochner, A., Faggart, M., et al. (2002) Science 296, 2225-2229. - PubMed
-
- Kruglyak, L. & Nickerson, D. A. (2001) Nat. Genet. 27, 234-236. - PubMed
-
- Morris, R. W. & Kaplan, N. L. (2002) Genet. Epidemiol. 23, 221-233. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

