Pharmacodynamic modeling of ciprofloxacin resistance in Staphylococcus aureus
- PMID: 15616298
- PMCID: PMC538881
- DOI: 10.1128/AAC.49.1.209-219.2005
Pharmacodynamic modeling of ciprofloxacin resistance in Staphylococcus aureus
Abstract
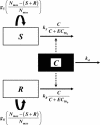
Three pharmacodynamic models of increasing complexity, designed for two subpopulations of bacteria with different susceptibilities, were developed to describe and predict the evolution of resistance to ciprofloxacin in Staphylococcus aureus by using pharmacokinetic, viable count, subpopulation, and resistance mechanism data obtained from in vitro system experiments. A two-population model with unique growth and killing rate constants for the ciprofloxacin-susceptible and -resistant subpopulations best described the initial killing and subsequent regrowth patterns observed. The model correctly described the enrichment of subpopulations with low-level resistance in the parent cultures but did not identify a relationship between the time ciprofloxacin concentrations were in the mutant selection window (the interval between the MIC and the mutant prevention concentration) and the enrichment of these subpopulations. The model confirmed the importance of resistant variants to the emergence of resistance by successfully predicting that resistant subpopulations would not emerge when a low-density culture, with a low probability of mutants, was exposed to a clinical dosing regimen or when a high-density culture, with a higher probability of mutants, was exposed to a transient high initial concentration designed to rapidly eradicate low-level resistant grlA mutants. The model, however, did not predict or explain the origin of variants with higher levels of resistance that appeared and became the predominant subpopulation during some experiments or the persistence of susceptible bacteria in other experiments where resistance did not emerge. Continued evaluation of the present two-population pharmacodynamic model and development of alternative models is warranted.
Figures



References
-
- Andersson, D. I., and B. R. Levin. 1999. The biological costs of antibiotic resistance. Curr. Opin. Microbiol. 2:489-493. - PubMed
-
- Austin, D. J., N. J. White, and R. M. Anderson. 1998. The dynamics of drug action on the within-host population growth of infectious agents: melding pharmacodynamics and pathogen population dynamics. J. Theor. Biol. 194:313-339. - PubMed
-
- Baquero, F., M. C. Negri, M. I. Morosini, and J. Blazquez. 1998. Antibiotic-selective environments. Clin. Infect. Dis. 27(Suppl. 1):S5-S11. - PubMed
-
- Baquero, F., M. C. Negri, M. I. Morosini, and J. Blazquez. 1997. The antibiotic selective process: concentration-specific amplification of low-level resistant populations, p. 93-111. In S. B. Levy (ed.), Antibiotic resistance: origins, evolution, selection, and spread. Wiley, Chichester, United Kingdom. - PubMed
-
- Bauernfeind, A. 1997. Comparison of the antibacterial activities of the quinolones Bay 12-8039, gatifloxacin (AM 1155), trovafloxacin, clinafloxacin, levofloxacin and ciprofloxacin. J. Antimicrob. Chemother. 40:639-651. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

