The ancient origin of the complement system
- PMID: 15616573
- PMCID: PMC545819
- DOI: 10.1038/sj.emboj.7600533
The ancient origin of the complement system
Abstract
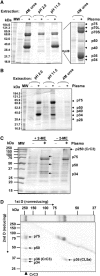
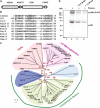
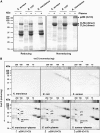
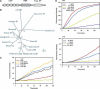
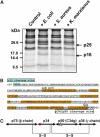
The complement system has been thought to originate exclusively in the deuterostomes. Here, we show that the central complement components already existed in the primitive protostome lineage. A functional homolog of vertebrate complement 3, CrC3, has been isolated from a 'living fossil', the horseshoe crab (Carcinoscorpius rotundicauda). CrC3 resembles human C3 and shows closest homology to C3 sequences of lower deuterostomes. CrC3 and plasma lectins bind a wide range of microbes, forming the frontline innate immune defense system. Additionally, we identified CrC2/Bf, a homolog of vertebrate C2 and Bf that participates in C3 activation, and a C3 receptor-like sequence. Furthermore, complement-mediated phagocytosis of bacteria by the hemocytes of horseshoe crab was also observed. Thus, a primitive yet complex opsonic complement defense system is revealed in the horseshoe crab, a protostome species. Our findings demonstrate an ancient origin of the critical complement components and the opsonic defense mechanism in the Precambrian ancestor of bilateral animals.
Figures


References
-
- Armstrong PB, Melchior R, Swarnakar S, Quigley JP (1998) Alpha2-macroglobulin does not function as a C3 homologue in the plasma hemolytic system of the American horseshoe crab, Limulus. Mol Immunol 35: 47–53 - PubMed
-
- Azumi K, De Santis R, De Tomaso A, Rigoutsos I, Yoshizaki F, Pinto MR, Marino R, Shida K, Ikeda M, Ikeda M, Arai M, Inoue Y, Shimizu T, Satoh N, Rokhsar DS, Du Pasquier L, Kasahara M, Satake M, Nonaka M (2003) Genomic analysis of immunity in a Urochordate and the emergence of the vertebrate immune system: ‘waiting for Godot'. Immunogenetics 55: 570–581 - PubMed
-
- Beutler B (2004) Innate immunity: an overview. Mol Immune 40: 845–859 - PubMed
-
- Chen SC, Yen CH, Yeh MS, Huang CJ, Liu TY (2001) Biochemical properties and cDNA cloning of two new lectins from the plasma of Tachypleus tridentatus: Tachypleus plasma lectin 1 and 2+. J Biol Chem 276: 9631–9639 - PubMed
-
- Davis AE III, Harrison RA, Lachmann PJ (1984) Physiologic inactivation of fluid phase C3b: isolation and structural analysis of C3c, C3d,g (alpha 2D), and C3g. J Immunol 132: 1960–1966 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

