The VirE3 protein of Agrobacterium mimics a host cell function required for plant genetic transformation
- PMID: 15616576
- PMCID: PMC545813
- DOI: 10.1038/sj.emboj.7600524
The VirE3 protein of Agrobacterium mimics a host cell function required for plant genetic transformation
Abstract
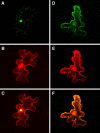
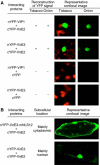
To genetically transform plants, Agrobacterium exports its transferred DNA (T-DNA) and several virulence (Vir) proteins into the host cell. Among these proteins, VirE3 is the only one whose biological function is completely unknown. Here, we demonstrate that VirE3 is transferred from Agrobacterium to the plant cell and then imported into its nucleus via the karyopherin alpha-dependent pathway. In addition to binding plant karyopherin alpha, VirE3 interacts with VirE2, a major bacterial protein that directly associates with the T-DNA and facilitates its nuclear import. The VirE2 nuclear import in turn is mediated by a plant protein, VIP1. Our data indicate that VirE3 can mimic this VIP1 function, acting as an 'adapter' molecule between VirE2 and karyopherin alpha and 'piggy-backing' VirE2 into the host cell nucleus. As VIP1 is not an abundant protein, representing one of the limiting factors for transformation, Agrobacterium may have evolved to produce and export to the host cells its own virulence protein that at least partially complements the cellular VIP1 function necessary for the T-DNA nuclear import and subsequent expression within the infected cell.
Figures






References
-
- Bartel P, Chien C, Sternglanz R, Fields S (1993) Elimination of false positives that arise in using the two-hybrid system. BioTechniques 14: 920–924 - PubMed
-
- Braem CV, Kas K, Meyen E, Debiec-Rychter M, Van De Ven WJ, Voz ML (2002) Identification of a karyopherin alpha 2 recognition site in PLAG1, which functions as a nuclear localization signal. J Biol Chem 277: 19673–19678 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

