Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance
- PMID: 15616582
- PMCID: PMC545806
- DOI: 10.1038/sj.emboj.7600511
Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance
Abstract
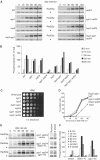
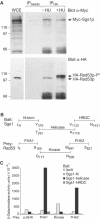
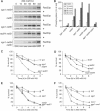
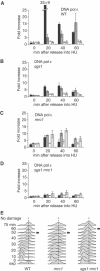
The RecQ helicase Sgs1p forms a complex with the type 1 DNA topoisomerase Top3p that resolves double Holliday junctions resulting from Rad51-mediated exchange. We find, however, that Sgs1p functions independently of both Top3p and Rad51p to stimulate the checkpoint kinase Rad53p when replication forks stall due to dNTP depletion on hydroxyurea. Checkpoint activation does not require Sgs1p function as a helicase, and correlates with its ability to bind the Rad53p kinase FHA1 motif directly. On the other hand, Sgs1p's helicase activity is required together with Top3p and the strand-exchange factor Rad51p, to help stabilise DNA polymerase epsilon at stalled replication forks. In this function, the Sgs1p/Top3p complex acts in parallel to the Claspin-related adaptor, Mrc1p, although the sgs1 and mrc1 mutations are epistatic for Rad53p activation. We thus identify two distinct pathways through which Sgs1p contributes to genomic integrity: checkpoint kinase activation requires Sgs1p as a noncatalytic Rad53p-binding site, while the combined Top3p/Sgs1p resolvase activity contributes to replisome stability and recovery from arrested replication forks.
Figures


References
-
- Adams A, Gottschling DE, Kaiser CA, Stearns T (1997) Methods in Yeast Genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press
-
- Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ (2001) Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol 3: 958–965 - PubMed
-
- Aushubel FM, Brent R, Kinston R, Moore D, Seidman JJ, Smith J, Struhl K (1994) Current Protocols in Molecular Biology. New York: John Wiley and Sons
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

