Transcriptional adaptation of Shigella flexneri during infection of macrophages and epithelial cells: insights into the strategies of a cytosolic bacterial pathogen
- PMID: 15618144
- PMCID: PMC538992
- DOI: 10.1128/IAI.73.1.88-102.2005
Transcriptional adaptation of Shigella flexneri during infection of macrophages and epithelial cells: insights into the strategies of a cytosolic bacterial pathogen
Abstract
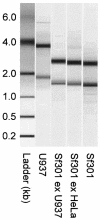
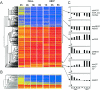
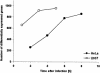
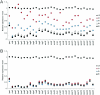
Shigella flexneri, the etiologic agent of bacillary dysentery, invades epithelial cells as well as macrophages and dendritic cells and escapes into the cytosol soon after invasion. Dissection of the global gene expression profile of the bacterium in its intracellular niche is essential to fully understand the biology of Shigella infection. We have determined the complete gene expression profiles for S. flexneri infecting human epithelial HeLa cells and human macrophage-like U937 cells. Approximately one quarter of the S. flexneri genes showed significant transcriptional adaptation during infection; 929 and 1,060 genes were up- or down-regulated within HeLa cells and U937 cells, respectively. The key S. flexneri virulence genes, ipa-mxi-spa and icsA, were drastically down-regulated during intracellular growth. This theme seems to be common in bacterial infection, because the Ipa-Mxi-Spa-like type III secretion systems were also down-regulated during mammalian cell infection by Salmonella enterica serovar Typhimurium and Escherichia coli O157. The bacteria experienced restricted levels of iron, magnesium, and phosphate in both host cell types, as shown by up-regulation of the sitABCD system, the mgtA gene, and genes of the phoBR regulon. Interestingly, ydeO and other acid-induced genes were up-regulated only in U937 cells and not in HeLa cells, suggesting that the cytosol of U937 cells is acidic. Comparison with the gene expression of intracellular Salmonella serovar Typhimurium, which resides within the Salmonella-containing vacuole, indicated that S. flexneri is exposed to oxidative stress in U937 cells. This work will facilitate functional studies of hundreds of novel intracellularly regulated genes that may be important for the survival and growth strategies of Shigella in the human host.
Figures



References
-
- Bartoleschi, C., M. C. Pardini, C. Scaringi, M. C. Martino, C. Pazzani, and M. L. Bernardini. 2002. Selection of Shigella flexneri candidate virulence genes specifically induced in bacteria resident in host cell cytoplasm. Cell. Microbiol. 4:613-626. - PubMed
-
- Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57: 289-300.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

