Identification of a nematode chemosensory gene family
- PMID: 15618405
- PMCID: PMC539308
- DOI: 10.1073/pnas.0408307102
Identification of a nematode chemosensory gene family
Abstract
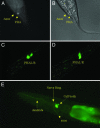
Taking advantage of the recent availability of the whole genome sequence of Caenorhabditis briggsae, a closely related nematode to Caenorhabditis elegans, we have examined the chemosensory gene superfamily by using comparative genomic methods. We have identified a chemosensory gene family, serpentine receptor class ab (srab), which exists in both species with 25 members in C. elegans and 14 members in C. briggsae. More than 20% of these gene models are reannotated. The srab family is similar to, but distinct from, the previously described serpentine receptor class a (sra) family and shows a differential expansion in C. elegans similar to that previously described for sra. The cellular expression patterns for multiple members of the srab family in both phasmid neurons in the tail and amphid neurons in the head supports the conclusion that they are chemosensory genes and suggests that they may play a role in integrating chemosensory inputs from both ends of the organism. The expansion of both the srab and sra gene families in C. elegans relative to C. briggsae is due to multiple rounds of tandem duplication and translocation of individual genes.
Figures



References
-
- Spehr, M., Gisselmann, G., Poplawski, A., Riffell, J. A., Wetzel, C. H., Zimmer, R. K. & Hatt, H. (2003) Science 299, 2054-2058. - PubMed
-
- Buck, L. B. (2000) Cell 100, 611-618. - PubMed
-
- Mombaerts, P. (2004) Nat. Rev. Neurosci. 5, 263-278. - PubMed
-
- Burr, A. H. (1985) Photochem. Photobiol. 41, 577-582. - PubMed
-
- Robertson, H. M. (1998) Genome Res. 8, 449-463. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

