Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins
- PMID: 15623580
- PMCID: PMC2171683
- DOI: 10.1083/jcb.200407182
Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins
Abstract
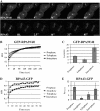
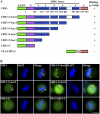
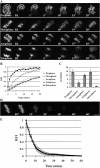
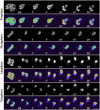
During mitosis, chromosomes are highly condensed and transcription is silenced globally. One explanation for transcriptional repression is the reduced accessibility of transcription factors. To directly test this hypothesis and to investigate the dynamics of mitotic chromatin, we evaluate the exchange kinetics of several RNA polymerase I transcription factors and nucleosome components on mitotic chromatin in living cells. We demonstrate that these factors rapidly exchange on and off ribosomal DNA clusters and that the kinetics of exchange varies at different phases of mitosis. In addition, the nucleosome component H1c-GFP also shows phase-specific exchange rates with mitotic chromatin. Furthermore, core histone components exchange at detectable levels that are elevated during anaphase and telophase, temporally correlating with H3-K9 acetylation and recruitment of RNA polymerase II before the onset of bulk RNA synthesis at mitotic exit. Our findings indicate that mitotic chromosomes in general and ribosomal genes in particular, although highly condensed, are accessible to transcription factors and chromatin proteins. The phase-specific exchanges of nucleosome components during late mitotic phases are consistent with an emerging model of replication independent core histone replacement.
Figures




References
-
- Ahmad, K., and S. Henikoff. 2002. a. Epigenetic consequences of nucleosome dynamics. Cell. 111:281–284. - PubMed
-
- Ahmad, K., and S. Henikoff. 2002. b. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 9:1191–1200. - PubMed
-
- Bell, S.P., R.M. Learned, H.M. Jantzen, and R. Tjian. 1988. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science. 241:1192–1197. - PubMed
-
- Belotserkovskaya, R., and D. Reinberg. 2004. Facts about FACT and transcript elongation through chromatin. Curr. Opin. Genet. Dev. 14:139–146. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

