Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis
- PMID: 15625105
- PMCID: PMC544298
- DOI: 10.1073/pnas.0408438102
Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis
Abstract
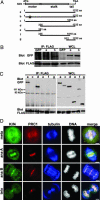
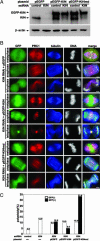
The spindle midzone, a conspicuous network of antiparallel interdigitating nonkinetochore microtubules between separating chromosomes, plays a crucial role in regulating the initiation and completion of cytokinesis. In this study, we report the use of time-lapse microscopy and a human kinesin endoribonucleases RNase III-prepared short interfering RNA (esiRNA) library to identify Kif4 as a motor protein that translocates PRC1, a spindle midzone-associated cyclin-dependent kinase substrate protein, to the plus ends of interdigitating spindle microtubules during the metaphase-to-anaphase transition. We show that Kif4 binds to PRC1 through its "stalk plus tail" domains and Kif4 and PRC1 colocalize on the spindle midzone/midbody during anaphase and cytokinesis. Suppression of Kif4 expression by Kif4 esiRNA results in the inhibition of PRC1 translocation, a block of the midzone formation, and a failure of cytokinesis. PRC1 translocation and midzone formation can be restored, and the cytokinetic defects can be rescued in Kif4 esiRNA-treated cells by coexpression of Kif4 but not its motor dead mutant Kif4md. Furthermore, we show that cyclin-dependent kinase phosphorylation of PRC1 controls the timing of PRC1 translocation by Kif4. These results, in light of the crucial role of PRC1 in midzone formation, indicate that cell cycle-dependent translocation of PRC1 by Kif4 is essential for midzone formation and cytokinesis.
Figures


Similar articles
-
Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation.EMBO J. 2004 Aug 18;23(16):3237-48. doi: 10.1038/sj.emboj.7600347. Epub 2004 Aug 5. EMBO J. 2004. PMID: 15297875 Free PMC article.
-
Spatiotemporal control of spindle midzone formation by PRC1 in human cells.Proc Natl Acad Sci U S A. 2006 Apr 18;103(16):6196-201. doi: 10.1073/pnas.0506926103. Epub 2006 Apr 7. Proc Natl Acad Sci U S A. 2006. PMID: 16603632 Free PMC article.
-
Kinesin superfamily protein member 4 (KIF4) is localized to midzone and midbody in dividing cells.Exp Mol Med. 2004 Feb 29;36(1):93-7. doi: 10.1038/emm.2004.13. Exp Mol Med. 2004. PMID: 15031677
-
Cell cycle control of spindle elongation.Cell Cycle. 2010 Mar 15;9(6):1084-90. doi: 10.4161/cc.9.6.11017. Epub 2010 Mar 15. Cell Cycle. 2010. PMID: 20410686 Review.
-
The role of centromere-binding factor 3 (CBF3) in spindle stability, cytokinesis, and kinetochore attachment.Biochem Cell Biol. 2005 Dec;83(6):696-702. doi: 10.1139/o05-161. Biochem Cell Biol. 2005. PMID: 16333320 Review.
Cited by
-
Midbody assembly and its regulation during cytokinesis.Mol Biol Cell. 2012 Mar;23(6):1024-34. doi: 10.1091/mbc.E11-08-0721. Epub 2012 Jan 25. Mol Biol Cell. 2012. PMID: 22278743 Free PMC article.
-
Classical and Emerging Regulatory Mechanisms of Cytokinesis in Animal Cells.Biology (Basel). 2019 Jul 26;8(3):55. doi: 10.3390/biology8030055. Biology (Basel). 2019. PMID: 31357447 Free PMC article. Review.
-
The ciliopathy protein CCDC66 controls mitotic progression and cytokinesis by promoting microtubule nucleation and organization.PLoS Biol. 2022 Jul 18;20(7):e3001708. doi: 10.1371/journal.pbio.3001708. eCollection 2022 Jul. PLoS Biol. 2022. PMID: 35849559 Free PMC article.
-
A novel PHD-finger protein 14/KIF4A complex overexpressed in lung cancer is involved in cell mitosis regulation and tumorigenesis.Oncotarget. 2017 Mar 21;8(12):19684-19698. doi: 10.18632/oncotarget.14962. Oncotarget. 2017. PMID: 28160558 Free PMC article.
-
New Auroras on the Roles of the Chromosomal Passenger Complex in Cytokinesis: Implications for Cancer Therapies.Front Oncol. 2015 Oct 14;5:221. doi: 10.3389/fonc.2015.00221. eCollection 2015. Front Oncol. 2015. PMID: 26528433 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

