Analysis on frequency and density of microsatellites in coding sequences of several eukaryotic genomes
- PMID: 15629040
- PMCID: PMC5172436
- DOI: 10.1016/s1672-0229(04)02004-2
Analysis on frequency and density of microsatellites in coding sequences of several eukaryotic genomes
Abstract
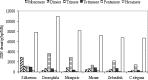
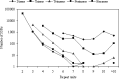
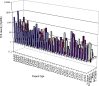
Microsatellites or simple sequence repeats (SSRs) have been found in most organisms during the last decade. Since large-scale sequences are being generated, especially those that can be used to search for microsatellites, the development of these markers is getting more convenient. Keeping SSRs in viewing the importance of the application, available CDS (coding sequences) or ESTs (expressed sequence tags) of some eukaryotic species were used to study the frequency and density of various types of microsatellites. On the basis of surveying CDS or EST sequences amounting to 66.6 Mb in silkworm, 37.2 Mb in fly, 20.8 Mb in mosquito, 60.0 Mb in mouse, 34.9 Mb in zebrafish and 33.5 Mb in Caenorhabditis elegans, the frequency of SSRs was 1/1.00 Kb in silkworm, 1/0.77 Kb in fly, 1/1.03 Kb in mosquito, 1/1.21 Kb in mouse, 1/1.25 Kb in zebrafish and 1/1.38 Kb in C. elegans. The overall average SSR frequency of these species is 1/1.07 Kb. Hexanucleotide repeats (64.5%-76.6%) are the most abundant class of SSR in the investigated species, followed by trimeric, dimeric, tetrameric, monomeric and pentameric repeats. Furthermore, the A-rich repeats are predominant in each type of SSRs, whereas G-rich repeats are rare in the coding regions.
Figures




References
-
- Dib C. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature. 1996;380:149–152. - PubMed
-
- Kashi Y. Simple sequence repeats as a source of quantitative genetic variation. Trends Genet. 1997;13:74–78. - PubMed
-
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Yu J. Inconsistency between SSR groupings and genetic backgrounds of white corn inbreds. Maydica. 2001;46:133–139.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

