The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants
- PMID: 15629062
- PMCID: PMC544883
- DOI: 10.1186/1471-2148-5-1
The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants
Abstract
Background: WRKY proteins are newly identified transcription factors involved in many plant processes including plant responses to biotic and abiotic stresses. To date, genes encoding WRKY proteins have been identified only from plants. Comprehensive search for WRKY genes in non-plant organisms and phylogenetic analysis would provide invaluable information about the origin and expansion of the WRKY family.
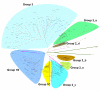
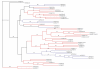
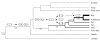
Results: We searched all publicly available sequence data for WRKY genes. A single copy of the WRKY gene encoding two WRKY domains was identified from Giardia lamblia, a primitive eukaryote, Dictyostelium discoideum, a slime mold closely related to the lineage of animals and fungi, and the green alga Chlamydomonas reinhardtii, an early branching of plants. This ancestral WRKY gene seems to have duplicated many times during the evolution of plants, resulting in a large family in evolutionarily advanced flowering plants. In rice, the WRKY gene family consists of over 100 members. Analyses suggest that the C-terminal domain of the two-WRKY-domain encoding gene appears to be the ancestor of the single-WRKY-domain encoding genes, and that the WRKY domains may be phylogenetically classified into five groups. We propose a model to explain the WRKY family's origin in eukaryotes and expansion in plants.
Conclusions: WRKY genes seem to have originated in early eukaryotes and greatly expanded in plants. The elucidation of the evolution and duplicative expansion of the WRKY genes should provide valuable information on their functions.
Figures
References
-
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu GL. Arabidopsis transcription factors: genome-wide comparative analysis among Eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

