Evolutionarily divergent extradiol dioxygenases possess higher specificities for polychlorinated biphenyl metabolites
- PMID: 15629912
- PMCID: PMC543568
- DOI: 10.1128/JB.187.2.415-421.2005
Evolutionarily divergent extradiol dioxygenases possess higher specificities for polychlorinated biphenyl metabolites
Abstract
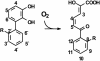
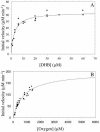
The reactivities of four evolutionarily divergent extradiol dioxygenases towards mono-, di-, and trichlorinated (triCl) 2,3-dihydroxybiphenyls (DHBs) were investigated: 2,3-dihydroxybiphenyl dioxygenase (EC 1.13.11.39) from Burkholderia sp. strain LB400 (DHBDLB400), DHBDP6-I and DHBDP6-III from Rhodococcus globerulus P6, and 2,2',3-trihydroxybiphenyl dioxygenase from Sphingomonas sp. strain RW1 (THBDRW1). The specificity of each isozyme for particular DHBs differed by up to 3 orders of magnitude. Interestingly, the Kmapp values of each isozyme for the tested polychlorinated DHBs were invariably lower than those of monochlorinated DHBs. Moreover, each enzyme cleaved at least one of the tested chlorinated (Cl) DHBs better than it cleaved DHB (e.g., apparent specificity constants for 3',5'-dichlorinated [diCl] DHB were 2 to 13.4 times higher than for DHB). These results are consistent with structural data and modeling studies which indicate that the substrate-binding pocket of the DHBDs is hydrophobic and can accommodate the Cl DHBs, particularly in the distal portion of the pocket. Although the activity of DHBDP6-III was generally lower than that of the other three enzymes, six of eight tested Cl DHBs were better substrates for DHBDP6-III than was DHB. Indeed, DHBDP6-III had the highest apparent specificity for 4,3',5'-triCl DHB and cleaved this compound better than two of the other enzymes. Of the four enzymes, THBDRW1 had the highest specificity for 2'-Cl DHB and was at least five times more resistant to inactivation by 2'-Cl DHB, consistent with the similarity between the latter and 2,2',3-trihydroxybiphenyl. Nonetheless, THBDRW1 had the lowest specificity for 2',6'-diCl DHB and, like the other enzymes, was unable to cleave this critical PCB metabolite (kcatapp < 0.001 s(-1)).
Figures
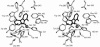
Similar articles
-
Characterization of extradiol dioxygenases from a polychlorinated biphenyl-degrading strain that possess higher specificities for chlorinated metabolites.J Bacteriol. 2003 Feb;185(4):1253-60. doi: 10.1128/JB.185.4.1253-1260.2003. J Bacteriol. 2003. PMID: 12562795 Free PMC article.
-
Dehalogenation, denitration, dehydroxylation, and angular attack on substituted biphenyls and related compounds by a biphenyl dioxygenase.J Bacteriol. 2001 Jun;183(12):3548-55. doi: 10.1128/JB.183.12.3548-3555.2001. J Bacteriol. 2001. PMID: 11371517 Free PMC article.
-
Substrate specificity and expression of three 2,3-dihydroxybiphenyl 1,2-dioxygenases from Rhodococcus globerulus strain P6.J Bacteriol. 2003 May;185(9):2944-51. doi: 10.1128/JB.185.9.2944-2951.2003. J Bacteriol. 2003. PMID: 12700274 Free PMC article.
-
[Molecular biology of enzymes degrading pollutants: diversity and convergent evolution observed on extradiol dioxygenases].Tanpakushitsu Kakusan Koso. 2000 Jun;45(8):1339-49. Tanpakushitsu Kakusan Koso. 2000. PMID: 10846472 Review. Japanese. No abstract available.
-
[PCB degradation systems in Rhodococcus bacteria: multiplex enzyme system determined by multiple isozyme genes].Tanpakushitsu Kakusan Koso. 2005 Oct;50(12):1541-7. Tanpakushitsu Kakusan Koso. 2005. PMID: 16218454 Review. Japanese. No abstract available.
Cited by
-
Surfing in the storm: how Paraburkholderia xenovorans thrives under stress during biodegradation of toxic aromatic compounds and other stressors.FEMS Microbiol Rev. 2025 Jan 14;49:fuaf021. doi: 10.1093/femsre/fuaf021. FEMS Microbiol Rev. 2025. PMID: 40388301 Free PMC article. Review.
-
Separate Upper Pathway Ring Cleavage Dioxygenases Are Required for Growth of Sphingomonas wittichii Strain RW1 on Dibenzofuran and Dibenzo-p-Dioxin.Appl Environ Microbiol. 2021 May 11;87(11):e02464-20. doi: 10.1128/AEM.02464-20. Print 2021 May 11. Appl Environ Microbiol. 2021. PMID: 33741618 Free PMC article.
-
Roles of ring-hydroxylating dioxygenases in styrene and benzene catabolism in Rhodococcus jostii RHA1.J Bacteriol. 2008 Jan;190(1):37-47. doi: 10.1128/JB.01122-07. Epub 2007 Oct 26. J Bacteriol. 2008. PMID: 17965160 Free PMC article.
-
A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages.Proc Natl Acad Sci U S A. 2007 Feb 6;104(6):1947-52. doi: 10.1073/pnas.0605728104. Epub 2007 Jan 30. Proc Natl Acad Sci U S A. 2007. PMID: 17264217 Free PMC article.
-
Probing the Mechanism of l‑DOPA 2,3-Dioxygenase Using Synthetic Derivatives of 3,4-Dihydroxyhydrocinnamic Acid.ACS Omega. 2025 Jul 16;10(29):32053-32069. doi: 10.1021/acsomega.5c03691. eCollection 2025 Jul 29. ACS Omega. 2025. PMID: 40757298 Free PMC article.
References
-
- Abramowicz, D. A. 1990. Aerobic and anaerobic biodegradation of PCBs: a review. Crit. Rev. Biotechnol. 10:241-251.
-
- Asturias, J. A., L. D. Eltis, M. Prucha, and K. N. Timmis. 1994. Analysis of three 2,3-dihydroxybiphenyl 1,2-dioxygenases found in Rhodococcus globerulus P6. J. Biol. Chem. 269:7807-7815. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2000. Current protocols in molecular biology. J. Wiley & Sons Inc., New York, N.Y.
-
- Barriault, D., M. Vedadi, J. Powlowski, and M. Sylvestre. 1999. cis-2,3-Dihydro-2,3-dihydroxybiphenyl dehydrogenase and cis-1,2-dihydro-1,2-dihydroxynaphthalene dehydrogenase catalyze dehydrogenation of the same range of substrates. Biochem. Biophys. Res. Commun. 260:181-187. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

