The bkdR gene of Streptomyces coelicolor is required for morphogenesis and antibiotic production and encodes a transcriptional regulator of a branched-chain amino acid dehydrogenase complex
- PMID: 15629937
- PMCID: PMC543559
- DOI: 10.1128/JB.187.2.664-671.2005
The bkdR gene of Streptomyces coelicolor is required for morphogenesis and antibiotic production and encodes a transcriptional regulator of a branched-chain amino acid dehydrogenase complex
Abstract
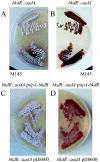
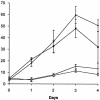
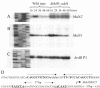
Products from the degradation of the branched-chain amino acids valine, leucine, and isoleucine contribute to the production of a number of important cellular metabolites, including branched-chain fatty acids, ATP and other energy production, cell-cell signaling for morphological development, and the synthesis of precursors for polyketide antibiotics. The first nonreversible reactions in the degradation of all three amino acids are catalyzed by the same branched-chain alpha-keto acid dehydrogenase (BCDH) complex. Actinomycetes are apparently unique among bacteria in that they contain two separate gene clusters, each of which encodes a BCDH enzyme complex. Here, we show that one of these clusters in Streptomyces coelicolor is regulated, at least in part, at the level of transcription by the product of the bkdR gene. The predicted product of this gene is a protein with similarity to a family of proteins that respond to leucine and serve to activate transcription of amino acid utilization operons. Unlike most other members of this class, however, the S. coelicolor bkdR gene product serves to repress transcription, suggesting that the branched-chain amino acids act as inducers rather than coactivators of transcription. BkdR likely responds to the presence of branched-chain amino acids. Its role in transcriptional regulation may be rationalized by the fact that transition from vegetative growth to aerial mycelium production, the first stage of morphological development in these complex bacteria, is coincident with extensive cellular lysis generating abundant amounts of protein that likely serve as the predominant source of carbon and nitrogen for metabolism. We suggest that bkdR plays a key role in the ability of Streptomyces species to sense nutrient availability and redirect metabolism for the utilization of branched-chain amino acids for energy, carbon, and perhaps even morphogen synthesis. A null mutant of bkdR is itself defective in morphogenesis and antibiotic production, suggesting that the role of the bkdR gene product may be more global than specific nutrient utilization.
Figures



Similar articles
-
Regulatory and biosynthetic effects of the bkd gene clusters on the production of daptomycin and its analogs A21978C1-3.J Ind Microbiol Biotechnol. 2018 Apr;45(4):271-279. doi: 10.1007/s10295-018-2011-y. Epub 2018 Feb 7. J Ind Microbiol Biotechnol. 2018. PMID: 29411202
-
In vitro transcriptional studies of the bkd operon of Pseudomonas putida: L-branched-chain amino acids and D-leucine are the inducers.J Bacteriol. 1999 May;181(9):2889-94. doi: 10.1128/JB.181.9.2889-2894.1999. J Bacteriol. 1999. PMID: 10217783 Free PMC article.
-
The bkdR gene of Pseudomonas putida is required for expression of the bkd operon and encodes a protein related to Lrp of Escherichia coli.J Bacteriol. 1993 Jul;175(13):3934-40. doi: 10.1128/jb.175.13.3934-3940.1993. J Bacteriol. 1993. PMID: 8320210 Free PMC article.
-
The Gene bldA, a regulator of morphological differentiation and antibiotic production in streptomyces.Arch Pharm (Weinheim). 2015 Jul;348(7):455-62. doi: 10.1002/ardp.201500073. Epub 2015 Apr 27. Arch Pharm (Weinheim). 2015. PMID: 25917027 Review.
-
RNA degradation and the regulation of antibiotic synthesis in Streptomyces.Future Microbiol. 2010 Mar;5(3):419-29. doi: 10.2217/fmb.10.14. Future Microbiol. 2010. PMID: 20210552 Review.
Cited by
-
Characterization and engineering of the Lrp/AsnC family regulator SACE_5717 for erythromycin overproduction in Saccharopolyspora erythraea.J Ind Microbiol Biotechnol. 2019 Jul;46(7):1013-1024. doi: 10.1007/s10295-019-02178-2. Epub 2019 Apr 23. J Ind Microbiol Biotechnol. 2019. PMID: 31016583
-
A new paradigm for the regulation of A40926B0 biosynthesis.Synth Syst Biotechnol. 2025 Apr 7;10(3):794-806. doi: 10.1016/j.synbio.2025.03.012. eCollection 2025 Sep. Synth Syst Biotechnol. 2025. PMID: 40297762 Free PMC article.
-
Branched-chain amino acid catabolism provides precursors for the Type II polyketide antibiotic, actinorhodin, via pathways that are nutrient dependent.J Ind Microbiol Biotechnol. 2009 Jan;36(1):129-37. doi: 10.1007/s10295-008-0480-0. Epub 2008 Oct 8. J Ind Microbiol Biotechnol. 2009. PMID: 18841403
-
Direct Monitoring of Membrane Fatty Acid Changes and Effects on the Isoleucine/Valine Pathways in an ndgR Deletion Mutant of Streptomyces coelicolor.J Microbiol Biotechnol. 2023 Jun 28;33(6):724-735. doi: 10.4014/jmb.2301.01016. Epub 2023 Mar 22. J Microbiol Biotechnol. 2023. PMID: 37072678 Free PMC article.
-
Origin of the 3-methylglutaryl moiety in caprazamycin biosynthesis.Microb Cell Fact. 2022 Nov 5;21(1):232. doi: 10.1186/s12934-022-01955-6. Microb Cell Fact. 2022. PMID: 36335365 Free PMC article.
References
-
- Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. - PubMed
-
- Brinkman, A. B., T. J. Ettema, W. M. de Vos, and J. van der Oost. 2003. The Lrp family of transcriptional regulators. Mol. Microbiol. 48:287-294. - PubMed
-
- Chater, K. F., C. J. Bruton, K. A. Plaskitt, M. J. Buttner, C. Mendez, and J. D. Helmann. 1989. The developmental fate of S. coelicolor hyphae depends upon a gene product homologous with the motility sigma factor of B. subtilis. Cell 59:133-143. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

