Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms
- PMID: 15630022
- PMCID: PMC540229
- DOI: 10.1101/gad.1262905
Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms
Abstract
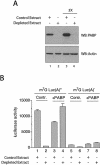
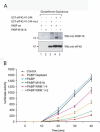
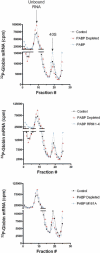
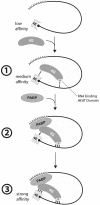
Translation initiation is a multistep process involving several canonical translation factors, which assemble at the 5'-end of the mRNA to promote the recruitment of the ribosome. Although the 3' poly(A) tail of eukaryotic mRNAs and its major bound protein, the poly(A)-binding protein (PABP), have been studied extensively, their mechanism of action in translation is not well understood and is confounded by differences between in vivo and in vitro systems. Here, we provide direct evidence for the involvement of PABP in key steps of the translation initiation pathway. Using a new technique to deplete PABP from mammalian cell extracts, we show that extracts lacking PABP exhibit dramatically reduced rates of translation, reduced efficiency of 48S and 80S ribosome initiation complex formation, and impaired interaction of eIF4E with the mRNA cap structure. Supplementing PABP-depleted extracts with wild-type PABP completely rectified these deficiencies, whereas a mutant of PABP, M161A, which is incapable of interacting with eIF4G, failed to restore translation. In addition, a stronger inhibition (approximately twofold) of 80S as compared to 48S ribosome complex formation (approximately 65% vs. approximately 35%, respectively) by PABP depletion suggests that PABP plays a direct role in 60S subunit joining. PABP can thus be considered a canonical translation initiation factor, integral to initiation complex formation at the 5'-end of mRNA.
Figures



References
-
- Afonina E., Neumann, M., and Pavlakis, G.N. 1997. Preferential binding of poly(A)-binding protein 1 to an inhibitory RNA element in the human immunodeficiency virus type 1 gag mRNA. J. Biol. Chem. 272: 2307–2311. - PubMed
-
- Bi X. and Goss, D.J. 2000. Wheat germ poly(A)-binding protein increases the ATPase and the RNA helicase activity of translation initiation factors eIF4A, eIF4B, and eIF-iso4F. J. Biol. Chem. 275: 17740–17746. - PubMed
-
- Edery I., Humbelin, M., Darveau, A., Lee, K.A., Milburn, S., Hershey, J.W., Trachsel, H., and Sonenberg, N. 1983. Involvement of eukaryotic initiation factor 4A in the cap recognition process. J. Biol. Chem. 258: 11398–11403. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
