Premature targeting of a cell division protein to midcell allows dissection of divisome assembly in Escherichia coli
- PMID: 15630023
- PMCID: PMC540231
- DOI: 10.1101/gad.1253805
Premature targeting of a cell division protein to midcell allows dissection of divisome assembly in Escherichia coli
Abstract
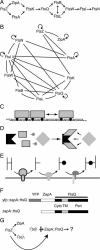
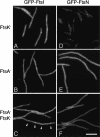
Cell division in Escherichia coli requires the recruitment of at least 10 essential proteins to the bacterial midcell. Recruitment of these proteins follows a largely linear dependency pathway in which depletion of one cell division protein leads to the absence from the division site of "downstream" proteins in the pathway. Analysis of events that underlie this pathway is complicated by the fact that a protein's ability to recruit "downstream" proteins is dependent on its own recruitment by "upstream" proteins. Hence, one cannot separate the individual contributions of various upstream proteins to any specific recruitment step. Here we present a method--premature targeting--for bypassing the normal localization requirements of a cell division protein and apply it to FtsQ, a protein recruited midway through the pathway. We fused FtsQ to the FtsZ-binding protein ZapA such that FtsQ was targeted to FtsZ rings independently of proteins FtsA and FtsK, which are normally required for FtsQ localization. Analysis of the resulting ZapA-FtsQ fusion suggests that FtsQ associates with a large complex of cell division proteins and that premature targeting of FtsQ can restore localization of this complex under conditions in which neither FtsQ nor the associated proteins would normally be localized.
Figures






Similar articles
-
Mutants, suppressors, and wrinkled colonies: mutant alleles of the cell division gene ftsQ point to functional domains in FtsQ and a role for domain 1C of FtsA in divisome assembly.J Bacteriol. 2007 Jan;189(2):633-45. doi: 10.1128/JB.00991-06. Epub 2006 Sep 15. J Bacteriol. 2007. PMID: 16980443 Free PMC article.
-
Premature targeting of cell division proteins to midcell reveals hierarchies of protein interactions involved in divisome assembly.Mol Microbiol. 2006 Jul;61(1):33-45. doi: 10.1111/j.1365-2958.2006.05206.x. Mol Microbiol. 2006. PMID: 16824093
-
Cell Cycle-Dependent Recruitment of FtsN to the Divisome in Escherichia coli.mBio. 2022 Aug 30;13(4):e0201722. doi: 10.1128/mbio.02017-22. Epub 2022 Aug 15. mBio. 2022. PMID: 35968943 Free PMC article.
-
Diverse paths to midcell: assembly of the bacterial cell division machinery.Curr Biol. 2005 Jul 12;15(13):R514-26. doi: 10.1016/j.cub.2005.06.038. Curr Biol. 2005. PMID: 16005287 Review.
-
Spatial control of the cell division site by the Min system in Escherichia coli.Environ Microbiol. 2013 Dec;15(12):3229-39. doi: 10.1111/1462-2920.12119. Epub 2013 Apr 9. Environ Microbiol. 2013. PMID: 23574354 Review.
Cited by
-
Divin: a small molecule inhibitor of bacterial divisome assembly.J Am Chem Soc. 2013 Jul 3;135(26):9768-76. doi: 10.1021/ja404640f. Epub 2013 Jun 18. J Am Chem Soc. 2013. PMID: 23738839 Free PMC article.
-
FtsZ-ZapA-ZapB interactome of Escherichia coli.J Bacteriol. 2012 Jan;194(2):292-302. doi: 10.1128/JB.05821-11. Epub 2011 Nov 4. J Bacteriol. 2012. PMID: 22056926 Free PMC article.
-
A role for FtsA in SPOR-independent localization of the essential Escherichia coli cell division protein FtsN.Mol Microbiol. 2014 Jun;92(6):1212-26. doi: 10.1111/mmi.12623. Epub 2014 May 8. Mol Microbiol. 2014. PMID: 24750258 Free PMC article.
-
The bypass of ZipA by overexpression of FtsN requires a previously unknown conserved FtsN motif essential for FtsA-FtsN interaction supporting a model in which FtsA monomers recruit late cell division proteins to the Z ring.Mol Microbiol. 2015 Mar;95(6):971-87. doi: 10.1111/mmi.12907. Epub 2015 Feb 4. Mol Microbiol. 2015. PMID: 25496259 Free PMC article.
-
Mycobacterium tuberculosis ftsZ expression and minimal promoter activity.Tuberculosis (Edinb). 2009 Dec;89 Suppl 1(Suppl 1):S60-4. doi: 10.1016/S1472-9792(09)70014-9. Tuberculosis (Edinb). 2009. PMID: 20006308 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
