Genetic analysis of the kinetochore DASH complex reveals an antagonistic relationship with the ras/protein kinase A pathway and a novel subunit required for Ask1 association
- PMID: 15632076
- PMCID: PMC543429
- DOI: 10.1128/MCB.25.2.767-778.2005
Genetic analysis of the kinetochore DASH complex reveals an antagonistic relationship with the ras/protein kinase A pathway and a novel subunit required for Ask1 association
Abstract
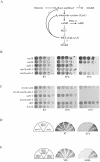
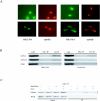
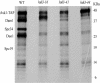
DASH is a microtubule- and kinetochore-associated complex required for proper chromosome segregation and bipolar attachment of sister chromatids on the mitotic spindle. We have undertaken a genetic and biochemical analysis of the DASH complex and uncovered a strong genetic interaction of DASH with the Ras/protein kinase A (PKA) pathway. Overexpression of PDE2 or deletion of RAS2 rescued the temperature sensitivity of ask1-3 mutants. Ras2 negatively regulates DASH through the PKA pathway. Constitutive PKA activity caused by mutation of the negative regulator BCY1 is toxic to DASH mutants such as ask1 and dam1. In addition, we have discovered two novel subunits of DASH, Hsk2 and Hsk3 (helper of Ask1), which are microproteins of fewer than 75 amino acids, as dosage suppressors of ask1 mutants. These are essential genes that colocalize with DASH components on spindles and kinetochores and are present in the DASH complex. Mutants in hsk3 arrest cells in mitosis with short spindles and broken spindle structures characteristic of other DASH mutants. Hsk3 is critical for the integrity of the DASH complex because in hsk3 mutants the association of Dam1, Duo1, Spc34, and Spc19 with Ask1 is greatly diminished. We propose that Hsk3 acts to incorporate Ask1 into the DASH complex.
Figures




Similar articles
-
The mitotic spindle is required for loading of the DASH complex onto the kinetochore.Genes Dev. 2002 Jan 15;16(2):183-97. doi: 10.1101/gad.959402. Genes Dev. 2002. PMID: 11799062 Free PMC article.
-
Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation.EMBO J. 2002 Jan 15;21(1-2):181-93. doi: 10.1093/emboj/21.1.181. EMBO J. 2002. PMID: 11782438 Free PMC article.
-
Glucose Signaling Is Connected to Chromosome Segregation Through Protein Kinase A Phosphorylation of the Dam1 Kinetochore Subunit in Saccharomycescerevisiae.Genetics. 2019 Feb;211(2):531-547. doi: 10.1534/genetics.118.301727. Epub 2018 Dec 13. Genetics. 2019. PMID: 30546002 Free PMC article.
-
Structures and functions of yeast kinetochore complexes.Annu Rev Biochem. 2007;76:563-91. doi: 10.1146/annurev.biochem.76.052705.160607. Annu Rev Biochem. 2007. PMID: 17362199 Review.
-
Structure-function insights into the yeast Dam1 kinetochore complex.J Cell Sci. 2009 Nov 1;122(Pt 21):3831-6. doi: 10.1242/jcs.004689. J Cell Sci. 2009. PMID: 19889968 Free PMC article. Review.
Cited by
-
The microprotein Nrs1 rewires the G1/S transcriptional machinery during nitrogen limitation in budding yeast.PLoS Biol. 2022 Mar 3;20(3):e3001548. doi: 10.1371/journal.pbio.3001548. eCollection 2022 Mar. PLoS Biol. 2022. PMID: 35239649 Free PMC article.
-
The composition, functions, and regulation of the budding yeast kinetochore.Genetics. 2013 Aug;194(4):817-46. doi: 10.1534/genetics.112.145276. Genetics. 2013. PMID: 23908374 Free PMC article. Review.
-
Genome-wide synthetic lethal screens identify an interaction between the nuclear envelope protein, Apq12p, and the kinetochore in Saccharomyces cerevisiae.Genetics. 2005 Oct;171(2):489-501. doi: 10.1534/genetics.105.045799. Epub 2005 Jul 5. Genetics. 2005. PMID: 15998715 Free PMC article.
-
Kinetochores require oligomerization of Dam1 complex to maintain microtubule attachments against tension and promote biorientation.Nat Commun. 2014 Sep 19;5:4951. doi: 10.1038/ncomms5951. Nat Commun. 2014. PMID: 25236177 Free PMC article.
-
Leveraging machine learning essentiality predictions and chemogenomic interactions to identify antifungal targets.Nat Commun. 2021 Nov 11;12(1):6497. doi: 10.1038/s41467-021-26850-3. Nat Commun. 2021. PMID: 34764269 Free PMC article.
References
-
- Adams, I. R., and J. V. Kilmartin. 2000. Spindle pole bodies duplication: a model for centrosome duplication? Trends Cell Biol. 10:329-335. - PubMed
-
- Amon, A. 1999. The spindle checkpoint. Curr. Opin. Genet. Dev. 9:69-75. - PubMed
-
- Aparicio, O. M., D. M. Weinstein, and S. P. Bell. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: restriction of MCM proteins and Cdc45p during S phase. Cell 91:59-69. - PubMed
-
- Biggins, S., and C. E. Walczak. 2003. Captivating capture: how microtubules attach to kinetochores. Curr. Biol. 13:R449-R460. - PubMed
-
- Bischoff, J. R., and G. D. Plowman. 1999. The Aurora/Ipl1p kinase family: regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 9:454-459. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
