Prss16 is not required for T-cell development
- PMID: 15632078
- PMCID: PMC543420
- DOI: 10.1128/MCB.25.2.789-796.2005
Prss16 is not required for T-cell development
Abstract
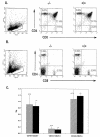
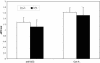
PRSS16 is a serine protease expressed exclusively in cortical thymic epithelial cells (cTEC) of the thymus, suggesting that it plays a role in the processing of peptide antigens during the positive selection of T cells. Moreover, the human PRSS16 gene is encoded in a region near the class I major histocompatibility complex (MHC) that has been linked to type 1 diabetes mellitus susceptibility. The mouse orthologue Prss16 is conserved in genetic structure, sequence, and pattern of expression. To study the role of Prss16 in thymic development, we generated a deletion mutant of Prss16 and characterized T-lymphocyte populations and MHC class II expression on cortical thymic epithelial cells. Prss16-deficient mice develop normally, are fertile, and show normal thymic morphology, cellularity, and anatomy. The total numbers and frequencies of thymocytes and splenic T-cell populations did not differ from those of wild-type controls. Surface expression of MHC class II on cTEC was also similar in homozygous mutant and wild-type animals, and invariant chain degradation was not impaired by deletion of Prss16. These findings suggest that Prss16 is not required for quantitatively normal T-cell development.
Figures
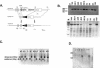


Similar articles
-
Thymus-specific serine protease regulates positive selection of a subset of CD4+ thymocytes.Eur J Immunol. 2009 Apr;39(4):956-64. doi: 10.1002/eji.200839175. Eur J Immunol. 2009. PMID: 19283781
-
Transcription profiling of Prss16 (Tssp) can be used to find additional peptidase genes that are candidates for self-peptide generation in the thymus.Mol Biol Rep. 2012 Apr;39(4):4051-8. doi: 10.1007/s11033-011-1186-3. Epub 2011 Jul 20. Mol Biol Rep. 2012. PMID: 21773946
-
Cloning of a novel MHC-encoded serine peptidase highly expressed by cortical epithelial cells of the thymus.Cell Immunol. 1999 Sep 15;196(2):80-6. doi: 10.1006/cimm.1999.1543. Cell Immunol. 1999. PMID: 10527559
-
The adaptive phenotype of cortical thymic epithelial cells.Eur J Immunol. 2009 Apr;39(4):944-7. doi: 10.1002/eji.200939315. Eur J Immunol. 2009. PMID: 19350578
-
Expression, genomic structure and mapping of the thymus specific protease prss16: a candidate gene for insulin dependent diabetes mellitus susceptibility.J Autoimmun. 2002 Jun;18(4):311-6. doi: 10.1006/jaut.2002.0593. J Autoimmun. 2002. PMID: 12144812
Cited by
-
Thymus-specific serine protease controls autoreactive CD4 T cell development and autoimmune diabetes in mice.J Clin Invest. 2011 May;121(5):1810-21. doi: 10.1172/JCI43314. Epub 2011 Apr 18. J Clin Invest. 2011. PMID: 21505262 Free PMC article.
-
Thymus-specific serine protease contributes to the diversification of the functional endogenous CD4 T cell receptor repertoire.J Exp Med. 2011 Jan 17;208(1):3-11. doi: 10.1084/jem.20100027. Epub 2010 Dec 20. J Exp Med. 2011. PMID: 21173102 Free PMC article.
-
Antigen presentation in the thymus for positive selection and central tolerance induction.Nat Rev Immunol. 2009 Dec;9(12):833-44. doi: 10.1038/nri2669. Nat Rev Immunol. 2009. PMID: 19935803 Review.
-
Thymus-specific serine protease, a protease that shapes the CD4 T cell repertoire.Immunogenetics. 2019 Mar;71(3):223-232. doi: 10.1007/s00251-018-1078-y. Epub 2018 Sep 17. Immunogenetics. 2019. PMID: 30225612 Review.
-
Gene expression profiling of precursor T-cell lymphoblastic leukemia/lymphoma identifies oncogenic pathways that are potential therapeutic targets.Leukemia. 2007 Jun;21(6):1276-84. doi: 10.1038/sj.leu.2404685. Epub 2007 Apr 12. Leukemia. 2007. PMID: 17429429 Free PMC article.
References
-
- Anderson, G., and E. J. Jenkinson. 2001. Lymphostromal interactions in thymic development and function. Nat. Rev. Immunol. 1:31-40. - PubMed
-
- Arnold, P. Y., N. L. La Gruta, T. Miller, K. M. Vignali, P. S. Adams, D. L. Woodland, and D. A. Vignali. 2002. The majority of immunogenic epitopes generate CD4+ T cells that are dependent on MHC class II-bound peptide-flanking residues. J. Immunol. 169:739-749. - PubMed
-
- Baldwin, K. K., B. P. Trenchak, J. D. Altman, and M. M. Davis. 1999. Negative selection of T cells occurs throughout thymic development. J. Immunol. 163:689-698. - PubMed
-
- Barton, G. M., and A. Y. Rudensky. 1999. Requirement for diverse, low-abundance peptides in positive selection of T cells. Science 283:67-70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
