The histone chaperone TAF-I/SET/INHAT is required for transcription in vitro of chromatin templates
- PMID: 15632079
- PMCID: PMC543418
- DOI: 10.1128/MCB.25.2.797-807.2005
The histone chaperone TAF-I/SET/INHAT is required for transcription in vitro of chromatin templates
Abstract
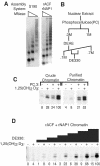
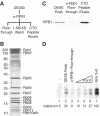
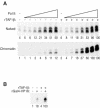
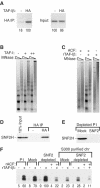
To uncover factors required for transcription by RNA polymerase II on chromatin, we fractionated a mammalian cell nuclear extract. We identified the histone chaperone TAF-I (also known as INHAT [inhibitor of histone acetyltransferase]), which was previously proposed to repress transcription, as a potent activator of chromatin transcription responsive to the vitamin D3 receptor or to Gal4-VP16. TAF-I associates with chromatin in vitro and can substitute for the related protein NAP-1 in assembling chromatin onto cloned DNA templates in cooperation with the remodeling enzyme ATP-dependent chromatin assembly factor (ACF). The chromatin assembly and transcriptional activation functions are distinct, however, and can be dissociated temporally. Efficient transcription of chromatin assembled with TAF-I still requires the presence of TAF-I during the polymerization reaction. Conversely, TAF-I cannot stimulate transcript elongation when added after the other factors necessary for assembly of a preinitiation complex on naked DNA. Thus, TAF-I is required to facilitate transcription at a step after chromatin assembly but before transcript elongation.
Figures



Similar articles
-
Herpes simplex virus type 1 tegument protein VP22 interacts with TAF-I proteins and inhibits nucleosome assembly but not regulation of histone acetylation by INHAT.J Gen Virol. 2003 Sep;84(Pt 9):2501-2510. doi: 10.1099/vir.0.19326-0. J Gen Virol. 2003. PMID: 12917472
-
Thanatos-associated protein 7 associates with template activating factor-Ibeta and inhibits histone acetylation to repress transcription.Mol Endocrinol. 2006 Feb;20(2):335-47. doi: 10.1210/me.2005-0248. Epub 2005 Sep 29. Mol Endocrinol. 2006. PMID: 16195249
-
Histone acetylation-independent transcription stimulation by a histone chaperone.Nucleic Acids Res. 2007;35(3):705-15. doi: 10.1093/nar/gkl1077. Epub 2006 Dec 19. Nucleic Acids Res. 2007. PMID: 17179179 Free PMC article.
-
Negotiating the nucleosome: factors that allow RNA polymerase II to elongate through chromatin.Biochem Cell Biol. 2007 Aug;85(4):426-34. doi: 10.1139/O07-054. Biochem Cell Biol. 2007. PMID: 17713578 Review.
-
Anp32e (Cpd1) and related protein phosphatase 2 inhibitors.Cerebellum. 2003;2(4):310-20. doi: 10.1080/14734220310017212. Cerebellum. 2003. PMID: 14964690 Review.
Cited by
-
Mitochondrial cytochrome c shot towards histone chaperone condensates in the nucleus.FEBS Open Bio. 2021 Sep;11(9):2418-2440. doi: 10.1002/2211-5463.13176. Epub 2021 May 19. FEBS Open Bio. 2021. PMID: 33938164 Free PMC article. Review.
-
Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II.Nat Struct Mol Biol. 2012 Nov;19(11):1108-15. doi: 10.1038/nsmb.2399. Epub 2012 Oct 14. Nat Struct Mol Biol. 2012. PMID: 23064645 Free PMC article.
-
Targeting inhibitor 2 of protein phosphatase 2A as a therapeutic strategy for prostate cancer treatment.Cancer Biol Ther. 2013 Oct 1;14(10):962-72. doi: 10.4161/cbt.25943. Epub 2013 Aug 5. Cancer Biol Ther. 2013. PMID: 24025258 Free PMC article.
-
Activated glucocorticoid receptor interacts with the INHAT component Set/TAF-Ibeta and releases it from a glucocorticoid-responsive gene promoter, relieving repression: implications for the pathogenesis of glucocorticoid resistance in acute undifferentiated leukemia with Set-Can translocation.Mol Cell Endocrinol. 2008 Feb 13;283(1-2):19-31. doi: 10.1016/j.mce.2007.10.014. Epub 2007 Nov 17. Mol Cell Endocrinol. 2008. PMID: 18096310 Free PMC article.
-
Transcription regulation of the rRNA gene by a multifunctional nucleolar protein, B23/nucleophosmin, through its histone chaperone activity.Mol Cell Biol. 2008 May;28(10):3114-26. doi: 10.1128/MCB.02078-07. Epub 2008 Mar 10. Mol Cell Biol. 2008. PMID: 18332108 Free PMC article.
References
-
- Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14:657-666. - PubMed
-
- Akey, C. W., and K. Luger. 2003. Histone chaperones and nucleosome assembly. Curr. Opin. Struct. Biol. 13:6-14. - PubMed
-
- Belotserkovskaya, R., S. Oh, V. A. Bondarenko, G. Orphanides, V. M. Studitsky, and D. Reinberg. 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301:1090-1093. - PubMed
-
- Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2004. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell 14:667-673. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
