Alternative mRNA splicing of SMRT creates functional diversity by generating corepressor isoforms with different affinities for different nuclear receptors
- PMID: 15632172
- PMCID: PMC2720035
- DOI: 10.1074/jbc.M411514200
Alternative mRNA splicing of SMRT creates functional diversity by generating corepressor isoforms with different affinities for different nuclear receptors
Abstract
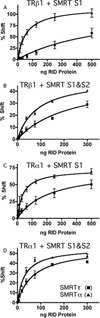
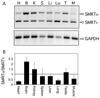
Many eukaryotic transcription factors are bimodal in their regulatory properties and can both repress and activate expression of their target genes. These divergent transcriptional properties are conferred through recruitment of auxiliary proteins, denoted coactivators and corepressors. Repression plays a particularly critical role in the functions of the nuclear receptors, a large family of ligand-regulated transcription factors involved in metazoan development, differentiation, reproduction, and homeostasis. The SMRT corepressor interacts directly with nuclear receptors and serves, in turn, as a platform for the assembly of a larger corepressor complex. We report here that SMRT is expressed in cells by alternative mRNA splicing to yield two distinct variants or isoforms. We designate these isoforms SMRTalpha and SMRTtau and demonstrate that these isoforms have significantly different affinities for different nuclear receptors. These isoforms are evolutionarily conserved and are expressed in a tissue-specific manner. Our results suggest that differential mRNA splicing serves to customize corepressor function in different cells, allowing the transcriptional properties of nuclear receptors to be adapted to different contexts.
Figures







Similar articles
-
Response of SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) and N-CoR (nuclear receptor corepressor) corepressors to mitogen-activated protein kinase kinase kinase cascades is determined by alternative mRNA splicing.Mol Endocrinol. 2007 Aug;21(8):1924-39. doi: 10.1210/me.2007-0035. Epub 2007 May 22. Mol Endocrinol. 2007. PMID: 17519355 Free PMC article.
-
Transcriptional silencing is defined by isoform- and heterodimer-specific interactions between nuclear hormone receptors and corepressors.Mol Cell Biol. 1998 Oct;18(10):5724-33. doi: 10.1128/MCB.18.10.5724. Mol Cell Biol. 1998. PMID: 9742089 Free PMC article.
-
Alternative splicing generates multiple SMRT transcripts encoding conserved repressor domains linked to variable transcription factor interaction domains.Nucleic Acids Res. 2004 Sep 1;32(15):4676-86. doi: 10.1093/nar/gkh786. Print 2004. Nucleic Acids Res. 2004. PMID: 15342788 Free PMC article.
-
Equilibrium interactions of corepressors and coactivators with agonist and antagonist complexes of glucocorticoid receptors.Mol Endocrinol. 2004 Jun;18(6):1376-95. doi: 10.1210/me.2003-0421. Epub 2004 Mar 11. Mol Endocrinol. 2004. PMID: 15016838
-
N-CoR-HDAC corepressor complexes: roles in transcriptional regulation by nuclear hormone receptors.Curr Top Microbiol Immunol. 2003;274:237-68. doi: 10.1007/978-3-642-55747-7_9. Curr Top Microbiol Immunol. 2003. PMID: 12596910 Review.
Cited by
-
Regulation of corepressor alternative mRNA splicing by hormonal and metabolic signaling.Mol Cell Endocrinol. 2015 Sep 15;413:228-35. doi: 10.1016/j.mce.2015.06.036. Epub 2015 Jul 10. Mol Cell Endocrinol. 2015. PMID: 26166430 Free PMC article.
-
Corepressors: custom tailoring and alterations while you wait.Nucl Recept Signal. 2005;3:e003. doi: 10.1621/nrs.03003. Epub 2005 Oct 21. Nucl Recept Signal. 2005. PMID: 16604171 Free PMC article.
-
Thyroid hormone receptor mutations in cancer and resistance to thyroid hormone: perspective and prognosis.J Thyroid Res. 2011;2011:361304. doi: 10.4061/2011/361304. Epub 2011 Jun 8. J Thyroid Res. 2011. PMID: 21760978 Free PMC article.
-
The actions of thyroid hormone signaling in the nucleus.Mol Cell Endocrinol. 2017 Dec 15;458:127-135. doi: 10.1016/j.mce.2017.03.001. Epub 2017 Mar 10. Mol Cell Endocrinol. 2017. PMID: 28286327 Free PMC article. Review.
-
Sequence analysis and identification of new isoform of EP4 receptors in different atlantic salmon tissues (Salmo salar L.) and its role in PGE2 induced immunomodulation in vitro.PLoS One. 2015 Apr 2;10(3):e0120483. doi: 10.1371/journal.pone.0120483. eCollection 2015. PLoS One. 2015. PMID: 25837516 Free PMC article.
References
-
- Sucov HM, Evans RM. Mol. Neurobiol. 1995;10:169–184. - PubMed
-
- Yen PM. Physiol. Rev. 2001;81:1097–1142. - PubMed
-
- Tsai MJ, O’Malley BW. Annu. Rev. Biochem. 1994;63:451–486. - PubMed
-
- Zhang J, Lazar MA. Annu. Rev. Physiol. 2000;62:439–466. - PubMed
-
- Kastner P, Mark M, Chambon P. Cell. 1995;83:859–869. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

