The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes
- PMID: 15634773
- PMCID: PMC6725199
- DOI: 10.1523/JNEUROSCI.3571-04.2005
The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes
Abstract
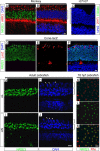
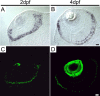
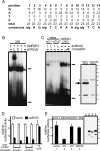
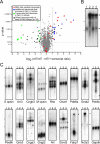
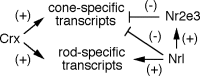
This study addresses one genetic regulatory mechanism that establishes the distinct identities of rod and cone photoreceptors. Previous work has shown that mutations in either humans or mice in the gene coding for photoreceptor-specific nuclear receptor Nr2e3 cause a progressive retinal degeneration characterized by increased numbers of short-wave cones. In the present work, we have examined the cellular and developmental pattern of Nr2e3 protein localization in mammals and fish, identified an optimal Nr2e3 DNA-binding site using cycles of binding to recombinant Nr2e3, characterized the transcriptional activity of wild type and one of the disease-associated point mutations in Nr2e3 in transfected cells, and characterized the transcriptional defects in the naturally occurring Nr2e3 mutant (rd7) mouse. These experiments indicate that in the mature vertebrate retina Nr2e3 is expressed exclusively in rods, that expression of Nr2e3 is one of the earliest events in the pathway of rod-specific photoreceptor development, and that Nr2e3 functions, either directly or indirectly, as a repressor of cone-specific genes in rod photoreceptor cells.
Figures



References
-
- Akhmedov NB, Piriev NI, Chang B, Rapoport AL, Hawes NL, Nishina PM, Nusinowitz S, Heckenlively JR, Roderick TH, Kozak CA, Danciger M, Davisson MT, Farber DB (2000) A deletion in a photoreceptor-specific nuclear receptor mRNA causes retinal degeneration in the rd7 mouse. Proc Natl Acad Sci USA 97: 5551-5556. - PMC - PubMed
-
- Anderson RE, Maude MB, Bok D (2001) Low docosahexaenoic acid levels in rod outer segment membranes of mice with rds/peripherin and P216L peripherin mutations. Invest Ophthalmol Vis Sci 42: 1715-1720. - PubMed
-
- Arannda A, Pascual A (2001) Nuclear hormone receptors and gene expression. Physiol Rev 81: 1269-1304. - PubMed
-
- Bessant DA, Payne AM, Mitton KP, Wang QL, Swain PK, Plant C, Bird AC, Zack DJ, Swaroop A, Bhattacharya SS (1999) A mutation in NRL is associated with autosomal dominant retinitis pigmentosa. Nat Genet 21: 355-356. - PubMed
-
- Bicknell IR, Darrow R, Barsalou L, Fliesler SJ, Organisciak DT (2002) Alterations in retinal rod outer segment fatty acids and light-damage susceptibility in P23H rats. Mol Vis 8: 333-340. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
