Evaluation of the COBAS TaqMan HCV test with automated sample processing using the MagNA pure LC instrument
- PMID: 15634985
- PMCID: PMC540141
- DOI: 10.1128/JCM.43.1.293-298.2005
Evaluation of the COBAS TaqMan HCV test with automated sample processing using the MagNA pure LC instrument
Abstract
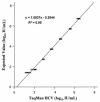
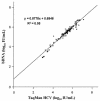
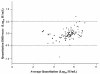
The COBAS TaqMan HCV Test (TaqMan HCV; Roche Molecular Systems Inc., Branchburg, N.J.) for hepatitis C virus (HCV) performed on the COBAS TaqMan 48 Analyzer (Roche Molecular Systems) currently relies on a manual sample processing method. Implementation of an automated sample processing method would facilitate the clinical use of this test. In this study, we evaluated the performance characteristics of TaqMan HCV following automated sample processing by the MagNA Pure LC instrument (MP; Roche Applied Science, Indianapolis, Ind.). The analytical sensitivity of TaqMan HCV following sample processing by MP was 8.1 IU/ml (95% confidence interval, 6.1 to 15.2). The assay showed good linearity (R(2) = 0.99) across a wide range of HCV RNA levels (25 to 5 x 10(6) IU/ml), with coefficients of variation ranging from 10% to 46%. Among 83 clinical specimens, the sensitivity and specificity of TaqMan HCV were 100% and 95%, respectively, when compared to the COBAS AMPLICOR hepatitis C virus test, version 2.0 (COBAS AMPLICOR; Roche Molecular Systems), with TaqMan HCV detecting two more HCV RNA-positive specimens than COBAS AMPLICOR. Both specimens were confirmed to be HCV RNA positive by the VERSANT HCV RNA qualitative test (Bayer HealthCare LLC, Tarrytown, N.Y.). There was also strong correlation (R(2) = 0.95) and good agreement between the results from TaqMan HCV and the VERSANT HCV RNA 3.0 assay (bDNA) (Bayer HealthCare LLC) among a group of 93 clinical specimens. The MP is a versatile, labor-saving sample processing platform suitable for reliable performance of TaqMan HCV.
Figures

References
-
- Albadalejo, J., R. Alonso, R. Antinozzi, M. Bogard, A. M. Bourgault, G. Colucci, T. Fenner, H. Petersen, E. Sala, J. Vincelette, and C. Young. 1998. Multicenter evaluation of the COBAS AMPLICOR HCV assay, an integrated PCR system for rapid detection of hepatitis C virus RNA in the diagnostic laboratory. J. Clin. Microbiol. 36:862-865. - PMC - PubMed
-
- Alter, M. J., D. Kruszon-Moran, O. V. Nainan, G. M. McQuillan, F. Gao, L. A. Moyer, R. A. Kaslow, and H. S. Margolis. 1999. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N. Engl. J. Med. 341:556-562. - PubMed
-
- Armstrong, G. L., M. J. Alter, G. M. McQuillan, and H. S. Margolis. 2000. The past incidence of hepatitis C virus infection: implications for the future burden of chronic liver disease in the United States. Hepatology 31:777-782. - PubMed
-
- Bland, J. M., and D. G. Altman. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet i:307-310. - PubMed
-
- Comanor, L., F. Anderson, M. Ghany, R. Perrillo, E. J. Heathcote, C. Sherlock, I. Zitron, D. Hendricks, and S. C. Gordon. 2001. Transcription-mediated amplification is more sensitive than conventional PCR-based assays for detecting residual serum HCV RNA at end of treatment. Am. J. Gastroenterol. 96:2968-2972. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

