Detection of human influenza A viruses by loop-mediated isothermal amplification
- PMID: 15635005
- PMCID: PMC540134
- DOI: 10.1128/JCM.43.1.427-430.2005
Detection of human influenza A viruses by loop-mediated isothermal amplification
Abstract
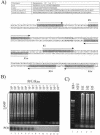
Here we describe the use of the loop-mediated isothermal amplification (LAMP) method to detect human influenza viruses (H1 to H3). Our results were correlated 100% with results deduced from routine clinical diagnostic tests. In addition, we also developed a LAMP assay specific for human beta-actin cDNA as a quality control test.
Figures

References
-
- Ellis, J. S., and M. C. Zambon. 2002. Molecular diagnosis of influenza. Rev. Med. Virol. 12:375-389. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

