High-resolution mapping of genotype-phenotype relationships in cri du chat syndrome using array comparative genomic hybridization
- PMID: 15635506
- PMCID: PMC1196376
- DOI: 10.1086/427762
High-resolution mapping of genotype-phenotype relationships in cri du chat syndrome using array comparative genomic hybridization
Abstract
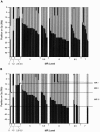
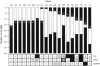
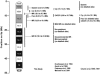
We have used array comparative genomic hybridization to map DNA copy-number changes in 94 patients with cri du chat syndrome who had been carefully evaluated for the presence of the characteristic cry, speech delay, facial dysmorphology, and level of mental retardation (MR). Most subjects had simple deletions involving 5p (67 terminal and 12 interstitial). Genotype-phenotype correlations localized the region associated with the cry to 1.5 Mb in distal 5p15.31, between bacterial artificial chromosomes (BACs) containing markers D5S2054 and D5S676; speech delay to 3.2 Mb in 5p15.32-15.33, between BACs containing D5S417 and D5S635; and the region associated with facial dysmorphology to 2.4 Mb in 5p15.2-15.31, between BACs containing D5S208 and D5S2887. These results overlap and refine those reported in previous publications. MR depended approximately on the 5p deletion size and location, but there were many cases in which the retardation was disproportionately severe, given the 5p deletion. All 15 of these cases, approximately two-thirds of the severely retarded patients, were found to have copy-number aberrations in addition to the 5p deletion. Restriction of consideration to patients with only 5p deletions clarified the effect of such deletions and suggested the presence of three regions, MRI-III, with differing effect on retardation. Deletions including MRI, a 1.2-Mb region overlapping the previously defined cri du chat critical region but not including MRII and MRIII, produced a moderate level of retardation. Deletions restricted to MRII, located just proximal to MRI, produced a milder level of retardation, whereas deletions restricted to the still-more proximal MRIII produced no discernible phenotype. However, MR increased as deletions that included MRI extended progressively into MRII and MRIII, and MR became profound when all three regions were deleted.
Figures

References
Electronic-Database Information
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for cri du chat syndrome)
-
- UCSC Genome Bioinformatics, http://genome.cse.ucsc.edu/
References
-
- Albertson DG, Pinkel D (2003) Genomic microarrays in human genetic disease and cancer. Hum Mol Genet 12 (Spec 2):R145–152 - PubMed
-
- Church DM, Yang J, Bocian M, Shiang R, Wasmuth JJ (1997) A high-resolution physical and transcript map of the cri du chat region of human chromosome 5p. Genome Res 7:787–801 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

