IRP1 Ser-711 is a phosphorylation site, critical for regulation of RNA-binding and aconitase activities
- PMID: 15636585
- PMCID: PMC1186702
- DOI: 10.1042/BJ20041623
IRP1 Ser-711 is a phosphorylation site, critical for regulation of RNA-binding and aconitase activities
Abstract
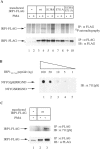
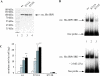
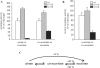
In iron-starved cells, IRP1 (iron regulatory protein 1) binds to mRNA iron-responsive elements and controls their translation or stability. In response to increased iron levels, RNA-binding is inhibited on assembly of a cubane [4Fe-4S] cluster, which renders IRP1 to a cytosolic aconitase. Phosphorylation at conserved serine residues may also regulate the activities of IRP1. We demonstrate that Ser-711 is a phosphorylation site in HEK-293 cells (human embryonic kidney 293 cells) treated with PMA, and we study the effects of the S711E (Ser-711-->Glu) mutation on IRP1 functions. A highly purified preparation of recombinant IRP1(S711E) displays negligible IRE-binding and aconitase activities. It appears that the first step in the aconitase reaction (conversion of citrate into the intermediate cis-aconitate) is more severely affected, as recombinant IRP1(S711E) retains approx. 45% of its capacity to catalyse the conversion of cis-aconitate into the end-product isocitrate. When expressed in mammalian cells, IRP1(S711E) completely fails to bind to RNA and to generate isocitrate from citrate. We demonstrate that the apparent inactivation of IRP1(S711E) is not related to mutation-associated protein misfolding or to alterations in its stability. Sequence analysis of IRP1 from all species currently deposited in protein databases shows that Ser-711 and flanking sequences are highly conserved in the evolutionary scale. Our results suggest that Ser-711 is a critical residue for the control of IRP1 activities.
Figures
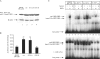


References
-
- Halliwell B., Gutteridge J. M. C. The role of free radicals and catalytic metal ions in human disease: an overview. Methods Enzymol. 1990;186:1–85. - PubMed
-
- Hentze M. W., Muckenthaler M. U., Andrews N. C. Balancing acts; molecular control of mammalian iron metabolism. Cell (Cambridge, Mass.) 2004;117:285–297. - PubMed
-
- Pantopoulos K. Iron metabolism and the IRE/IRP regulatory system: an update. Ann. N.Y. Acad. Sci. 2004;1012:1–13. - PubMed
-
- Ponka P., Beaumont C., Richardson D. R. Function and regulation of transferrin and ferritin. Semin. Hematol. 1998;35:35–54. - PubMed
-
- Eisenstein R. S., Tuazon P. T., Schalinske K. L., Anderson S. A., Traugh J. A. Iron-responsive element-binding protein. Phosphorylation by protein kinase C. J. Biol. Chem. 1993;268:27363–27370. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

