Linkage disequilibrium patterns and tagSNP transferability among European populations
- PMID: 15637659
- PMCID: PMC1196391
- DOI: 10.1086/427925
Linkage disequilibrium patterns and tagSNP transferability among European populations
Abstract
The pattern of linkage disequilibrium (LD) is critical for association studies, in which disease-causing variants are identified by allelic association with adjacent markers. The aim of this study is to compare the LD patterns in several distinct European populations. We analyzed four genomic regions (in total, 749 kb) containing candidate genes for complex traits. Individuals were genotyped for markers that are evenly distributed at an average spacing of approximately 2-4 kb in eight population-based samples from ongoing epidemiological studies across Europe. The Centre d'Etude du Polymorphisme Humain (CEPH) trios of the HapMap project were included and were used as a reference population. In general, we observed a conservation of the LD patterns across European samples. Nevertheless, shifts in the positions of the boundaries of high-LD regions can be demonstrated between populations, when assessed by a novel procedure based on bootstrapping. Transferability of LD information among populations was also tested. In two of the analyzed gene regions, sets of tagging single-nucleotide polymorphisms (tagSNPs) selected from the HapMap CEPH trios performed surprisingly well in all local European samples. However, significant variation in the other two gene regions predicts a restricted applicability of CEPH-derived tagging markers. Simulations based on our data set show the extent to which further gain in tagSNP efficiency and transferability can be achieved by increased SNP density.
Figures

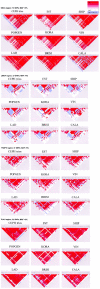






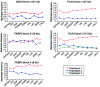
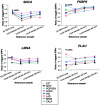

References
Electronic-Database Information
-
- GSF European LD Pattern Project, http://ihg.gsf.de/LD/ (for a downloadable version of the genotype data presented in this study)
-
- HapMap Homepage, http://www.hapmap.org/ (for the International HapMap Project)
-
- popgen, http://www.popgen.de/
References
-
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton, NJ
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

